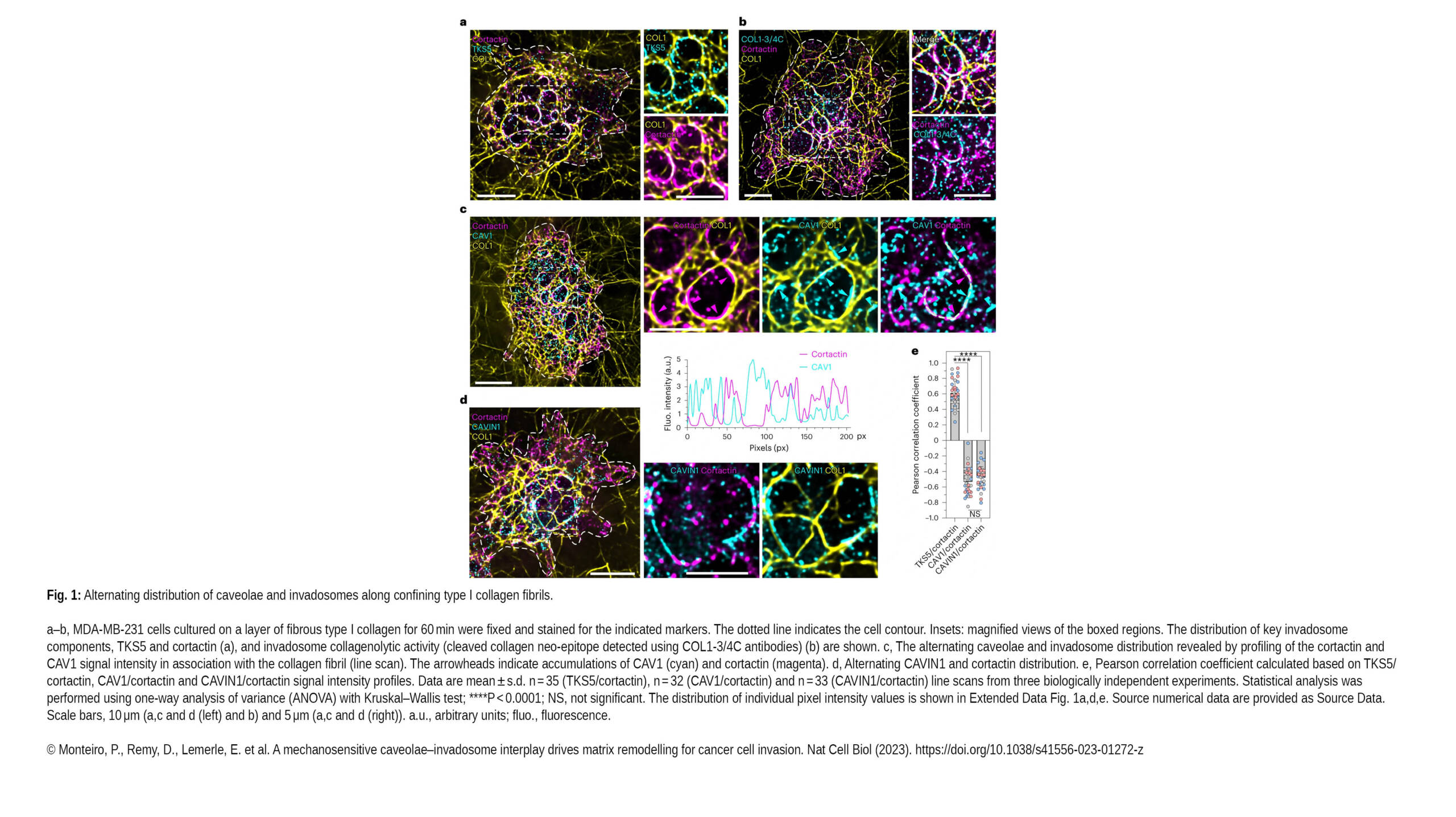
Le Ladoux / Mège recently contributed to the publication of a new article in Nature Cell biology :
A mechanosensitive caveolae–invadosome interplay drives matrix remodelling for cancer cell invasion
Abstract:
Invadosomes and caveolae are mechanosensitive structures that are implicated in metastasis. Here, we describe a unique juxtaposition of caveola clusters and matrix degradative invadosomes at contact…