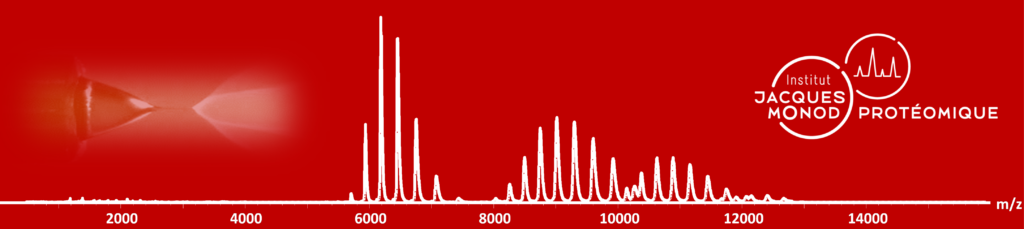
Proteomics: Mass Spectrometry
ProteoSeine is the proteomics facility of the Institut Jacques Monod, UMR 7592 Université de Paris Cité – CNRS. It has been located since early 2009 in the Buffon building at 15 rue Hélène Brion, 75205 Paris CEDEX 13 – France. The platform’s activities include analytical services, technical and methodological research and development, and active technology watch. In this context, it uses the most modern approaches of mass spectrometry for the study of protein constituents of living systems.
Its mission is to offer the biology research community state-of-the-art skills and services to integrate proteomic approaches into the development of their scientific projects.
ProteoSeine’s R&D is based on the active research in fundamental biology of the Institut Jacques Monod teams, on collaborations with the hospital sector and on a long-standing know-how in the analytical development of therapeutic proteins. The team is particularly developing its expertise in the high-throughput differential quantitative analysis of complex protein mixtures, the analysis of post-translational modifications, the characterisation of recombinant pharmaceutical proteins and the analysis of non-covalent complexes by mass spectrometry under native conditions. Bottom-up and top-down approaches at the nano or capillary scale are used according to the problems encountered.
The platform is labelled by the GIS IBiSA (Infrastructure for Biology, Health and Agronomy) and by the Cancéropôle Ile-de-France. It is open to the entire national and international, public and private scientific community. The traceability of activities is ensured through the registration of services and users in a LIMS specially developed internally and raw data are stored in redundancy via a solution managed globally by the Institut Jacques Monod. ProteoSeine is also an associate member of ProFi, the national research infrastructure dedicated to proteomic analysis and is supported by the following organisations:

ProteoSeine implements high-resolution electrospray mass spectrometers coupled to high-performance chromatographic systems.
Bottom-up analyses are performed on a latest generation timsTOF Pro2 (Bruker) coupled to a high-throughput separation instrument (Evosep One). The TIMS (Trapped Ion Mobility Spectrometry) technology is primarily a gas phase separation technique that solves sample complexity by adding an extra dimension of separation to high performance liquid chromatography and mass spectrometry. The TIMS device also accumulates and concentrates ions of a given m/z and mobility. By synchronising the ion accumulation and sequencing cycles in parallel (Parallel Accumulation-Serial Fragmentation (PASEF) technology), the device can offer extremely high MS/MS speed and sensitivity for in-depth proteome coverage and unbiased differential protein quantification.
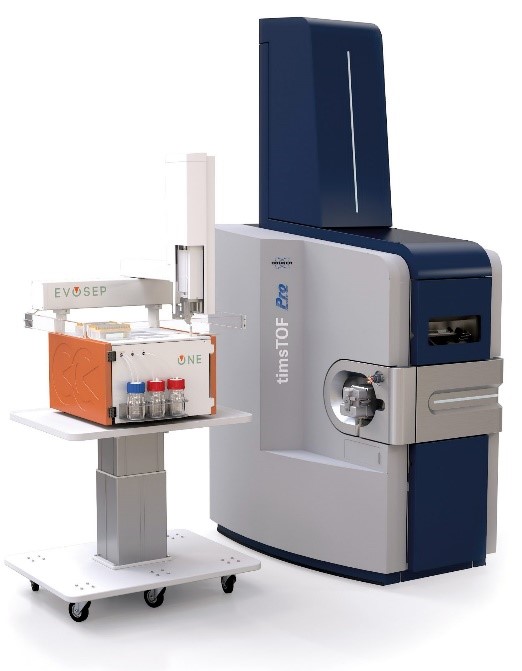
The platform is also equipped with proprietary software that allows reprocessing of raw data on two multi-core computing servers and multivariate statistical analysis of abundance results. The facility is in particularly good capacity to handle large sample sizes in automated workflows.
Native-MS and intact protein analyses are performed on a Cyclic IMS instrument (Waters) incorporating a revolutionary cyclic ion mobility technology. The instrument also has EXD and SID fragmentation capabilities in addition to the CID fragmentation mode available in the TRAP and TRANSFER cells. This instrument is coupled to a high-pressure capillary chromatographic system of the M-Class type or Acquity Premier for the development of high-resolution protein separations. An ADVION nanomate robot is also available for automated direct infusion analysis in nanospray mode.

ProteoSeine implements also a Hamilton liquid handler for automated sample preparation and a high-pressure chromatographic system for handling protein separations in different modalities.

The facility implements high-resolution electrospray mass spectrometers coupled to high-performance chromatographic systems. The instrumentation and bioinformatics tools used are specially selected to deliver state-of-the-art results in proteomic analysis.
Various analysis protocols are proposed by analysing intact proteins (top-down) or after digestion (bottom-up), which make it possible to tackle the following problems in particular:
- Identification of proteins on gel or in solution
- Characterisation of post-translational modifications (PTMs)
- Characterisation of recombinant proteins (coverage, PTMs, process & product-related impurities, size or charge variants)
- Search for partners (Pull-Down)
- Biomarker research
- Label-free quantitative analysis of proteome abundance variations
- Analysis of specific proteomes after enrichment (e.g. phosphoproteome)
- Analysis of complexes by MS under native conditions
The principle of operation is to first contact the platform to define an analysis protocol adapted to the project. A user sheet specifying how the platform works will be sent (also available in the “documents” section). Prior to any analysis, this form must be returned completed with the requested information. In a second step, it is sufficient to make an appointment to drop off the samples from Tuesday to Thursday in order for us to be able to process them as soon as possible.
The results generated are sent by e-mail within 2 weeks for classic approaches and are archived. The raw data are transmitted on demand as their storage and integrity are ensured by the platform in a non-contractual way. After receiving the results, the user is therefore responsible for the integrity of all his data.
The pricing of the facility’s services is calculated on the basis of full costs according to the CNRS DSFIM-SBOR-D-2014-46 reference system. Three distinct rates are applied according to the origin of the issuer of the order form, i.e. CNRS/UPC, other academic and private. The rates below are expressed exclusive of tax; they are to be increased by VAT at the current.
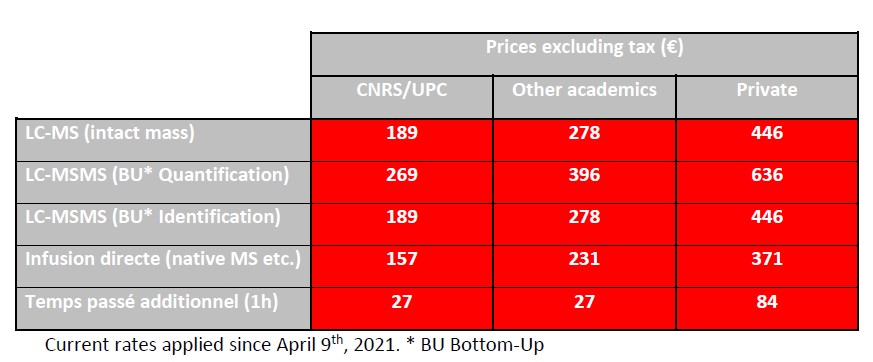
The mixed UPC/INSERM units under global INSERM management delegation listed below, nevertheless benefit from the CNRS/UPC rate.
The ProteoSeine team is heavily involved in initial and continuing education activities.
Each year, it provides a week of theoretical and practical training in proteomic analysis by mass spectrometry to students in the M2-Pro “Platform Engineering” at Université Paris Cité.
Every year, it hosts an M2-level student for a 6-month practical end-of-studies internship.
It organises training in mass spectrometry in quantitative proteomics and characterisation of recombinant proteins. This training is listed in the CNRS Formation Entreprise catalogue.
It organises two international workshops per year as part of the European Twinning H2020 programme “MiCoBion”.
To contact the facility:
spectrodemasse@ijm.fr, +33 (0)1 57 27 81 82
Team leader
 Guillaume CHEVREUX, ProtéoSeine proteomics facility manager, PFT/PROTEOSEINE+33 (0)1 57 27 81 19, room 657B
Guillaume CHEVREUX, ProtéoSeine proteomics facility manager, PFT/PROTEOSEINE+33 (0)1 57 27 81 19, room 657B
Members
 Jean-Baptiste BOYER, Proteomics engineer, PFT/PROTEOSEINE+33 (0)1 57 27 81 82, room 657B
Jean-Baptiste BOYER, Proteomics engineer, PFT/PROTEOSEINE+33 (0)1 57 27 81 82, room 657B Jean-Michel CAMADRO, Emeritus researcher, PFT/PROTEOSEINE+33 (0)1 57 27 80 95, room 344B
Jean-Michel CAMADRO, Emeritus researcher, PFT/PROTEOSEINE+33 (0)1 57 27 80 95, room 344B Victor COCHARD, Proteomics engineer, PFT/PROTEOSEINE+33 (0)1 57 27 81 82, room 657B
Victor COCHARD, Proteomics engineer, PFT/PROTEOSEINE+33 (0)1 57 27 81 82, room 657B Veronique LEGROS, Proteomics engineer, PFT/PROTEOSEINE+33 (0)1 57 27 81 82, room 657B
Veronique LEGROS, Proteomics engineer, PFT/PROTEOSEINE+33 (0)1 57 27 81 82, room 657B
To contact a member of the team by e-mail: name.surname@ijm.fr
ProteoSeine facility of Institut Jacques Monod, Bâtiment Buffon – Aile B, Pièce 657B, 6e étage – 15 rue Hélène Brion – 75205 Paris Cedex 13 – FRANCE
User’s charter
4202101_Fiche Utilisateur ProteoSeine_2022
Minimising contamination for proteomic analysis
Minimizing contamination in Proteomics
Cell lysis protocols for proteomic analysis
Publications in co-authorship with members of the facility (2017-2023)
2023
Lignieres L, Sénécaut N, Dang T, Bellutti L, Hamon M, Terrier S, Legros V, Chevreux G, Lelandais G, Mège RM, Dumont J, Camadro JM. Extending the Range of SLIM-Labeling Applications: From Human Cell Lines in Culture to Caenorhabditis elegans Whole-Organism Labeling. J Proteome Res. 2023 Mar 3;22(3):996-1002. doi: 10.1021/acs.jproteome.2c00699. Epub 2023 Feb 6. PMID: 36748112; PMCID: PMC9990122.
kombo Nkoula S, Velez-Aguilera G, Ossareh-Nazari B, Van Hove L, Ayuso C, Legros V, Chevreux G, Thomas L, Seydoux G, Askjaer P, Pintard L. Mechanisms of nuclear pore complex disassembly by the mitotic Polo-like kinase 1 (PLK-1) in C. elegans embryos. Sci Adv. 2023 Jul 21;9(29):eadf7826. doi: 10.1126/sciadv.adf7826. Epub 2023 Jul 19. PMID: 37467327; PMCID: PMC10355831.
Trouillard O, Dupaigne P, Dunoyer M, Doulazmi M, Herlin MK, Frismand S, Riou A, Legros V, Chevreux G, Veaute X, Busso D, Fouquet C, Saint-Martin C, Méneret A, Trembleau A, Dusart I, Dubacq C, Roze E. Congenital mirror movements are associated with defective polymerisation of RAD51. J Med Genet. 2023 Jun 12:jmg-2023-109189. doi: 10.1136/jmg-2023-109189. PMID: 37308287.
Nashed S, El Barbry H, Benchouaia M, Dijoux-Maréchal A, Delaveau T, Ruiz-Gutierrez N, Gaulier L, Tribouillard-Tanvier D, Chevreux G, Le Crom S, Palancade B, Devaux F, Laine E, Garcia M. Functional mapping of N-terminal residues in the yeast proteome uncovers novel determinants for mitochondrial protein import. PLoS Genet. 2023 Aug 16;19(8):e1010848. doi: 10.1371/journal.pgen.1010848. PMID: 37585488.
Gareil N, Gervais A, Macaisne N, Chevreux G, Canman JC, Andreani J, Dumont J. An unconventional TOG domain is required for CLASP localization. Curr Biol. 2023 Jul 21:S0960-9822(23)00916-8. doi: 10.1016/j.cub.2023.07.009. PMID: 37516114.
Zheng Y, Cabassa-Hourton C, Eubel H, Chevreux G, Lignieres L, Crilat E, Braun HP, Lebreton S, Savouré A. Pyrroline-5-carboxylate metabolism protein complex detected in Arabidopsis thaliana leaf mitochondria. J Exp Bot. 2023 Oct 16:erad406. doi: 10.1093/jxb/erad406. PMID: 37843921.
Postic, G., Solarz, J., Loubière, C., Kandiah, J., Sawmynaden, J., Adam, F., Vilaire, M., Léger, T., Camadro, J.-M., Victorino, D. B., Potier, M.-C., Bun, E., Moroy, G., Kauskot, A., Christophe, O., & Janel, N. (2023). Over-expression of Dyrk1A affects bleeding by modulating plasma fibronectin and fibrinogen level in mice. Journal of Cellular and Molecular Medicine, n/a(n/a). https://doi.org/10.1111/jcmm.17817
Scrima N, Le Bars R, Nevers Q, Glon D, Chevreux G, Civas A, Blondel D, Lagaudriere-Gesbert C, Gaudin Y (2023) Rabies virus P protein binds to TBK1 and interferes with the formation of innate immunity-related liquid condensates. Cell Rep. 42: 111949, doi: S2211-1247(22)01850-2, PMID: 36640307
Bayot J, Martin C, Chevreux G, Camadro JM, Auchere F (2023) The adaptive response to alternative carbon sources in the pathogen Candida albicans involves a remodeling of thiol-and glutathione-dependent redox status. Biochem J. doi: BCJ20220505, PMID: 36625375
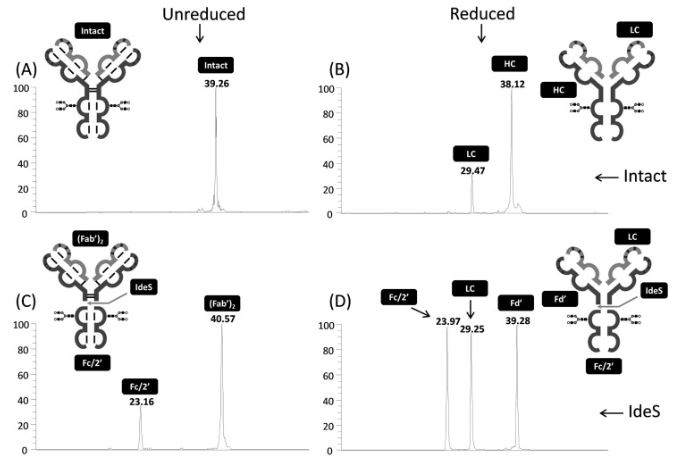
Lignieres L, Legros V, Khelil M, Senecaut N, Lauber MA, Camadro JM, Chevreux G (2023) Capillary liquid chromatography coupled with mass spectrometry for analysis of nanogram protein quantities on a wide-pore superficially porous particle column in top-down proteomics. J Chromatogr B Analyt Technol Biomed Life Sci. 1214: 123566, doi: S1570-0232(22)00471-8, PMID: 36516651
2022
Gallo G, Kotlik P, Roingeard P, Monot M, Chevreux G, Ulrich RG, Tordo N, Ermonval M (2022) Diverse susceptibilities and responses of human and rodent cells to orthohantavirus infection reveal different levels of cellular restriction. PLoS Negl Trop Dis. 16: e0010844, doi: 10.1371/journal.pntd.0010844, PMID: 36223391
Le Borgne P, Greibill L, Laporte MH, Lemullois M, Bouhouche K, Temagoult M, Rosnet O, Le Guennec M, Lignieres L, Chevreux G, Koll F, Hamel V, Guichard P, Tassin AM (2022) The evolutionary conserved proteins CEP90, FOPNL, and OFD1 recruit centriolar distal appendage proteins to initiate their assembly. PLoS Biol. 20: e3001782, doi: 10.1371/journal.pbio.3001782, PMID: 36070319
Senecaut N, Poulain P, Lignieres L, Terrier S, Legros V, Chevreux G, Lelandais G, Camadro JM (2022) Quantitative Proteomics in Yeast: From bSLIM and Proteome Discoverer Outputs to Graphical Assessment of the Significance of Protein Quantification Scores. Methods Mol Biol. 2477: 275-292, doi: 10.1007/978-1-0716-2257-5_16, PMID: 35524123
Miro-Pina C, Charmant O, Kawaguchi T, Holoch D, Michaud A, Cohen I, Humbert A, Jaszczyszyn Y, Chevreux G, Del Maestro L, Ait-Si-Ali S, Arnaiz O, Margueron R, Duharcourt S (2022) Paramecium Polycomb repressive complex 2 physically interacts with the small RNA-binding PIWI protein to repress transposable elements. Dev Cell. 57: 1037-1052 e1038, doi: 10.1016/j.devcel.2022.03.014, PMID: 35429435
Liu R, Zhang W, Gou P, Berthelet J, Nian Q, Chevreux G, Legros V, Moroy G, Bui LC, Wang L, Dupret JM, Deshayes F, Rodrigues Lima F (2022) Cisplatin causes covalent inhibition of protein-tyrosine phosphatase 1B (PTP1B) through reaction with its active site cysteine: Molecular, cellular and in vivo mice studies. Biomed Pharmacother. 153: 113372, doi: 10.1016/j.biopha.2022.113372, PMID: 35809481
Liu D, Marie JC, Pelletier AL, Song Z, Ben-Khemis M, Boudiaf K, Pintard C, Leger T, Terrier S, Chevreux G, El-Benna J, Dang PM (2022) Protein Kinase CK2 Acts as a Molecular Brake to Control NADPH Oxidase 1 Activation and Colon Inflammation. Cell Mol Gastroenterol Hepatol. 13: 1073-1093, doi: 10.1016/j.jcmgh.2022.01.003, PMID: 35031518
2021
Senecaut N, Alves G, Weisser H, Lignieres L, Terrier S, Yang-Crosson L, Poulain P, Lelandais G, Yu YK, Camadro JM (2021) Novel Insights into Quantitative Proteomics from an Innovative Bottom-Up Simple Light Isotope Metabolic (bSLIM) Labeling Data Processing Strategy. J Proteome Res. 20: 1476-1487, doi: 10.1021/acs.jproteome.0c00478, PMID: 33573382
Ribeiro-Parenti L, Jarry AC, Cavin JB, Willemetz A, Le Beyec J, Sannier A, Benadda S, Pelletier AL, Hourseau M, Leger T, Morlet B, Couvelard A, Anini Y, Msika S, Marmuse JP, Ledoux S, Le Gall M, Bado A (2021) Bariatric surgery induces a new gastric mucosa phenotype with increased functional glucagon-like peptide-1 expressing cells. Nat Commun. 12: 110, doi: 10.1038/s41467-020-20301-1, PMID: 33397977
Lemonnier T, Daldello EM, Poulhe R, Le T, Miot M, Lignieres L, Jessus C, Dupre A (2021) The M-phase regulatory phosphatase PP2A-B55delta opposes protein kinase A on Arpp19 to initiate meiotic division. Nat Commun. 12: 1837, doi: 10.1038/s41467-021-22124-0, PMID: 33758202
Lautier O, Penzo A, Rouviere JO, Chevreux G, Collet L, Loiodice I, Taddei A, Devaux F, Collart MA, Palancade B (2021) Co-translational assembly and localized translation of nucleoporins in nuclear pore complex biogenesis. Mol Cell. 81: 2417-2427 e2415, doi: 10.1016/j.molcel.2021.03.030, PMID: 33838103
Gallo G, Caignard G, Badonnel K, Chevreux G, Terrier S, Szemiel A, Roman-Sosa G, Binder F, Gu Q, Da Silva Filipe A, Ulrich RG, Kohl A, Vitour D, Tordo N, Ermonval M (2021) Interactions of Viral Proteins from Pathogenic and Low or Non-Pathogenic Orthohantaviruses with Human Type I Interferon Signaling. Viruses. 13: doi: 10.3390/v13010140, PMID: 33478127
Darrigrand R, Pierson A, Rouillon M, Renko D, Boulpicante M, Bouyssie D, Mouton-Barbosa E, Marcoux J, Garcia C, Ghosh M, Alami M, Apcher S (2021) Isoginkgetin derivative IP2 enhances the adaptive immune response against tumor antigens. Commun Biol. 4: 269, doi: 10.1038/s42003-021-01801-2, PMID: 33649389
Bertini EV, Torres MA, Leger T, Garcia C, Hong KW, Chong TM, Castellanos de Figueroa LI, Chan KG, Dessaux Y, Camadro JM, Nieto-Penalver CG (2021) Insight in the quorum sensing-driven lifestyle of the non-pathogenic Agrobacterium tumefaciens 6N2 and the interactions with the yeast Meyerozyma guilliermondii. Genomics. 113: 4352-4360, doi: 10.1016/j.ygeno.2021.11.017, PMID: 34793950
Balint B, Hergalant S, Camadro JM, Blaise S, Vanalderwiert L, Lignieres L, Gueant-Rodriguez RM, Gueant JL (2021) Fetal Programming by Methyl Donor Deficiency Produces Pathological Remodeling of the Ascending Aorta. Arterioscler Thromb Vasc Biol. 41: 1928-1941, doi: 10.1161/ATVBAHA.120.315587, PMID: 33827257
2020
Morilla I, Leger T, Marah A, Pic I, Zaag H, Ogier-Denis E (2020) Singular manifolds of proteomic drivers to model the evolution of inflammatory bowel disease status. Sci Rep. 10: 19066, doi: 10.1038/s41598-020-76011-7, PMID: 33149233
Duval C, Macabiou C, Garcia C, Lesuisse E, Camadro JM, Auchere F (2020) The adaptive response to iron involves changes in energetic strategies in the pathogen Candida albicans. Microbiologyopen. 9: e970, doi: 10.1002/mbo3.970, PMID: 31788966
2019
Scheiber IF, Pilatova J, Malych R, Kotabova E, Krijt M, Vyoral D, Mach J, Leger T, Camadro JM, Prasil O, Lesuisse E, Sutak R (2019) Copper and iron metabolism in Ostreococcus tauri – the role of phytotransferrin, plastocyanin and a chloroplast copper-transporting ATPase. Metallomics. 11: 1657-1666, doi: 10.1039/c9mt00078j, PMID: 31380866
Sanchez David RY, Combredet C, Najburg V, Millot GA, Beauclair G, Schwikowski B, Leger T, Camadro JM, Jacob Y, Bellalou J, Jouvenet N, Tangy F, Komarova AV (2019) LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses. Sci Signal. 12: doi: 10.1126/scisignal.aar3993, PMID: 31575732
Renaudin F, Sarda S, Campillo-Gimenez L, Severac C, Leger T, Charvillat C, Rey C, Liote F, Camadro JM, Ea HK, Combes C (2019) Adsorption of Proteins on m-CPPD and Urate Crystals Inhibits Crystal-induced Cell Responses: Study on Albumin-crystal Interaction. J Funct Biomater. 10: doi: 10.3390/jfb10020018, PMID: 31027151
Nian Q, Berthelet J, Zhang W, Bui LC, Liu R, Xu X, Duval R, Ganesan S, Leger T, Chomienne C, Busi F, Guidez F, Dupret JM, Rodrigues Lima F (2019) T-Cell Protein Tyrosine Phosphatase Is Irreversibly Inhibited by Etoposide-Quinone, a Reactive Metabolite of the Chemotherapy Drug Etoposide. Mol Pharmacol. 96: 297-306, doi: 10.1124/mol.119.116319, PMID: 31221825
Massarweh A, Bosco M, Chantret I, Leger T, Jamal L, Roper DI, Dowson CG, Busca P, Bouhss A, Gravier-Pelletier C, Moore SEH (2019) Bacterial Lipid II Analogs: Novel In Vitro Substrates for Mammalian Oligosaccharyl Diphosphodolichol Diphosphatase (DLODP) Activities. Molecules. 24: doi: 10.3390/molecules24112135, PMID: 31174247
Lelandais G, Denecker T, Garcia C, Danila N, Leger T, Camadro JM (2019) Label-free quantitative proteomics in Candida yeast species: technical and biological replicates to assess data reproducibility. BMC Res Notes. 12: 470, doi: 10.1186/s13104-019-4505-8, PMID: 31370875
Le Faouder J, Gigante E, Leger T, Albuquerque M, Beaufrere A, Soubrane O, Dokmak S, Camadro JM, Cros J, Paradis V (2019) Proteomic Landscape of Cholangiocarcinomas Reveals Three Different Subgroups According to Their Localization and the Aspect of Non-Tumor Liver. Proteomics Clin Appl. 13: e1800128, doi: 10.1002/prca.201800128, PMID: 30520266
Kundlacz C, Pourcelot M, Fablet A, Amaral Da Silva Moraes R, Leger T, Morlet B, Viarouge C, Sailleau C, Turpaud M, Gorlier A, Breard E, Lecollinet S, van Rijn PA, Zientara S, Vitour D, Caignard G (2019) Novel Function of Bluetongue Virus NS3 Protein in Regulation of the MAPK/ERK Signaling Pathway. J Virol. 93: doi: 10.1128/JVI.00336-19, PMID: 31167915
Fernandez J, Machado AK, Lyonnais S, Chamontin C, Gartner K, Leger T, Henriquet C, Garcia C, Portilho DM, Pugniere M, Chaloin L, Muriaux D, Yamauchi Y, Blaise M, Nisole S, Arhel NJ (2019) Transportin-1 binds to the HIV-1 capsid via a nuclear localization signal and triggers uncoating. Nat Microbiol. 4: 1840-1850, doi: 10.1038/s41564-019-0575-6, PMID: 31611641
Elzaiat M, Herman L, Legois B, Leger T, Todeschini AL, Veitia RA (2019) High-throughput Exploration of the Network Dependent on AKT1 in Mouse Ovarian Granulosa Cells. Mol Cell Proteomics. 18: 1307-1319, doi: 10.1074/mcp.RA119.0014613, PMID: 30992313
El Banna N, Hatem E, Heneman-Masurel A, Leger T, Baille D, Vernis L, Garcia C, Martineau S, Dupuy C, Vagner S, Camadro JM, Huang ME (2019) Redox modifications of cysteine-containing proteins, cell cycle arrest and translation inhibition: Involvement in vitamin C-induced breast cancer cell death. Redox Biol. 26: 101290, doi: 10.1016/j.redox.2019.101290, PMID: 31412312
Bouillier C, Cosentino G, Leger T, Rincheval V, Richard CA, Desquesnes A, Sitterlin D, Blouquit-Laye S, Eleouet JF, Gault E, Rameix-Welti MA (2019) The Interactome analysis of the Respiratory Syncytial Virus protein M2-1 suggests a new role in viral mRNA metabolism post-transcription. Sci Rep. 9: 15258, doi: 10.1038/s41598-019-51746-0, PMID: 31649314
Bonnet J, Garcia C, Leger T, Couquet MP, Vignoles P, Vatunga G, Ndung’u J, Boudot C, Bisser S, Courtioux B (2019) Proteome characterization in various biological fluids of Trypanosoma brucei gambiense-infected subjects. J Proteomics. 196: 150-161, doi: 10.1016/j.jprot.2018.11.005, PMID: 30414516
2018
Telot L, Rousseau E, Lesuisse E, Garcia C, Morlet B, Leger T, Camadro JM, Serre V (2018) Quantitative proteomics in Friedreich’s ataxia B-lymphocytes: A valuable approach to decipher the biochemical events responsible for pathogenesis. Biochim Biophys Acta Mol Basis Dis. 1864: 997-1009, doi: 10.1016/j.bbadis.2018.01.010, PMID: 29329987
Obino D, Fetler L, Soza A, Malbec O, Saez JJ, Labarca M, Oyanadel C, Del Valle Batalla F, Goles N, Chikina A, Lankar D, Segovia-Miranda F, Garcia C, Leger T, Gonzalez A, Espeli M, Lennon-Dumenil AM, Yuseff MI (2018) Galectin-8 Favors the Presentation of Surface-Tethered Antigens by Stabilizing the B Cell Immune Synapse. Cell Rep. 25: 3110-3122 e3116, doi: 10.1016/j.celrep.2018.11.052, PMID: 30540943
Denard J, Rouillon J, Leger T, Garcia C, Lambert MP, Griffith G, Jenny C, Camadro JM, Garcia L, Svinartchouk F (2018) AAV-8 and AAV-9 Vectors Cooperate with Serum Proteins Differently Than AAV-1 and AAV-6. Mol Ther Methods Clin Dev. 10: 291-302, doi: 10.1016/j.omtm.2018.08.001, PMID: 30155509
2017
Richarme G, Liu C, Mihoub M, Abdallah J, Leger T, Joly N, Liebart JC, Jurkunas UV, Nadal M, Bouloc P, Dairou J, Lamouri A (2017) Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science. 357: 208-211, doi: 10.1126/science.aag1095, PMID: 28596309
Planchon M, Leger T, Spalla O, Huber G, Ferrari R (2017) Metabolomic and proteomic investigations of impacts of titanium dioxide nanoparticles on Escherichia coli. PLoS One. 12: e0178437, doi: 10.1371/journal.pone.0178437, PMID: 28570583
Leger T, Garcia C, Collomb L, Camadro JM (2017) A Simple Light Isotope Metabolic Labeling (SLIM-labeling) Strategy: A Powerful Tool to Address the Dynamics of Proteome Variations In Vivo. Mol Cell Proteomics. 16: 2017-2031, doi: 10.1074/mcp.M117.066936, PMID: 28821603
Hovsepian J, Defenouillere Q, Albanese V, Vachova L, Garcia C, Palkova Z, Leon S (2017) Multilevel regulation of an alpha-arrestin by glucose depletion controls hexose transporter endocytosis. J Cell Biol. 216: 1811-1831, doi: 10.1083/jcb.201610094, PMID: 28468835
Gergondey R, Garcia C, Marchand CH, Lemaire SD, Camadro JM, Auchere F (2017) Modulation of the specific glutathionylation of mitochondrial proteins in the yeast Saccharomyces cerevisiae under basal and stress conditions. Biochem J. 474: 1175-1193, doi: 10.1042/BCJ20160927, PMID: 28167699
Boukpessi T, Hoac B, Coyac BR, Leger T, Garcia C, Wicart P, Whyte MP, Glorieux FH, Linglart A, Chaussain C, McKee MD (2017) Osteopontin and the dento-osseous pathobiology of X-linked hypophosphatemia. Bone. 95: 151-161, doi: 10.1016/j.bone.2016.11.019, PMID: 27884786
15/06/2023 :
The ProtéoSeine platform is organizing a seminar in conjunction with Protein Metrics. The aim of this seminar will be to present the work carried out by the platform, in particular on the analysis of intact proteins.
Protein Metrics will present its software solutions for protein, peptide and oligonucleotide analysis.
The seminar will take place on the morning of Thursday June 15, 2023, at the Institut Jacques Monod.
Would you like to take part?
You can view the program and register here :
https://proteinmetrics.com/paris-symposium-2023/
06/02/2023
Jean-Michel Camadro’s team, in collaboration with the ProtéoSeine platform, the Ladoux/Mège team and Dumont, have just published a new article:
Laurent Lignières, Nicolas Sénécaut, Tien Dang, Laura Bellutti, Marion Hamon, Samuel Terrier, Véronique Legros, Guillaume Chevreux, Gaëlle Lelandais, René-Marc Mège, Julien Dumont, and Jean-Michel Camadro, Journal of Proteome Research, 2023, 22, 3, 996–1002
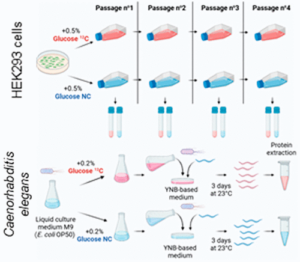
07/02/2023
Jean-Michel Camadro’s team in collaboration with the ProtéoSeine platform have just published a new article:
Juliette Bayot, Caroline Martin, Guillaume Chevreux, Jean-Michel Camadro, Françoise Auchère, Biochemical Journal Volume 480, Issue 3.
05/01/2023: the Protoseine platform has just published a new article in Journal of Chromatography B :
29/11/2022 : Inauguration de nouveaux équipements lors des 40 ans de l’IJM avec la Région Ile-de-France