Epigenomics and Paleogenomics
THIERRY GRANGE & EVA-MARIA GEIGL

We study the evolution of genomes to deepen the understanding of the relationship between genotypes and phenotypes. Recent phenotypic adaptations are ideal to explore the underlying genomic evolution while minimizing the confounding effects of genetic drift. We address these aspects through the analysis of ancient genomes from fossils, the direct witnesses of evolution, thus adding a geological or historical time-scale to evolutionary studies. We document evolutionary processes occurring over the last hundred thousand of years and push methodological limits to enable the study of more ancient samples from geographic regions where the hot climate is particularly detrimental to DNA preservation.
Keywords : evolution, genomes, ancient DNA, paleogenomics, peopling, domestication
+33 (0)1 57 27 81 29 / +33 (0)1 57 27 81 32 Contact Thierry GRANGE / Contact Eva-Maria GEIGL @thierrygrange.bsky.social
We focus on two major research lines: 1) Animal domestication, which represents a unique model of accelerated and human-driven evolution of specific genomic regions;2) Genomic evolution of human populations during the peopling of Europe over the last 40,000 years. These lines give rise to five interdisciplinary and interconnected research themes.
1.1. Paleogenomic study of the past population dynamics of bovines influenced by climate and humans.
Through the analysis of mitochondrial and nuclear genomes in up to 120,000-year-old fossils, we retrace migrations, contractions, expansions, extinctions and replacements of aurochs and bison populations. We aim to discriminate between environmental and human-induced effects on the evolution of these large herbivores by comparing two sister-species, Bos and Bison, as only Bos (cattle) was domesticated.
We reconstructed the phylogeographic evolution of bison, showed the influence of environmental conditions on the spread and replacement of lineages as well as sex-specific behavior at the origin of the discordance between mitochondrial lineages, morphotypes and modern genomes, and identified an unknown ancestor of the present-day European bison, which colonized Europe after the Last Glacial maximum (LGM) ~14,000 years ago (Massilani et al., 2016, BMC Biology; Grange et al., 2018, Diversity).
We have characterized the mitochondrial diversity of aurochs over a large sample size of 125 individuals using sequence capture of whole mitogenomes, and of more than 300 ancient individuals through PCR-based mitotype genotyping. This allowed us to follow the evolution of wild aurochs and domesticated cattle through time and space in Europe, south-west Asia and north Africa and to create an extended evolutionary framework as a response to both climate changes in the last 50,000 years and interaction with humans over the last 10,000 years (publication in prep.).
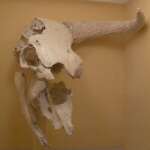
1.2. Paleogenomics of equids.
Three extant equid species live in the northern hemisphere, i.e., horses, African and Asiatic wild asses. While horses and African wild asses (donkeys) were domesticated and are ubiquitous, Asiatic wild asses (hemiones) were not and are on the edge of extinction. We identified paleogenetically the timing of the expansion of domestic horses (Guimaraes et al., 2020) and the population history of the hemiones (Bennett et al., 2017). The paleogenomic analysis of Bronze Age equid burials in Mesopotamia and the identification of extinct hemione populations allowed us to reconstruct the breeding techniques of the first equid hybrids known to date. These animals were used by the elite in warfare among Mesopotamian city-states in the 3rd millennium BCE (Bennett et al., 2022)
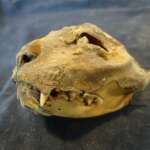
1.3. Paleogenomics of cat domestication.
Using ancient genomes, we attempt to reconstruct the domestication history of the cat over the last ~10,000 years, which is very different from the one of cattle. The cat’s domestication largely followed the commensal pathway and selective breeding became a prominent domestication mechanism only very recently. Using ancient mitochondrial DNA from archaeological specimens, we reconstructed the history of the spread of the north African/southwest Asian wildcat F.s.lybica, the predecessor of the domestic cat: it took place in two waves, the first one starting from the Fertile Crescent at the beginning of the Neolithic, the second from Egypt during Classical Antiquitys (Ottoni et al., 2017).
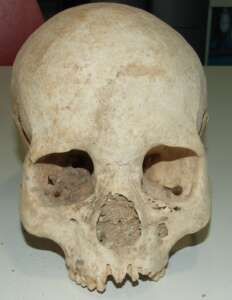
2.1. Morphology of Denisovans.
The discovery through paleogenomics of a hitherto unknown human population contemporary to the Neanderthals, the Denisovans, revolutionized paleoanthropology. Yet, the absence of morphologically characteristic skeletal remains of this population overshadowed this revolution. We analyzed the mitogenome of a distal finger bone and identified it as the Denisovan type specimen. It was at the time the only skeletal remain that could be morphologically described. The analysis performed by a collaborating paleoanthropologist showed that bone’s phenotypic traits are identical to those of Homo sapiens and that Neanderthals developed derived traits (Bennett et al., 2019).
2.2. Paleogenomics of the peopling of Europe.
We analyzed four important recent cultural and populational transition periods in European human history and explored the traces recorded in the genomes: 1) the period around 40,000 years ago when the first wave of Homo sapiens that had entered Europe experienced a dramatic environmental event, the Cambrian Ignimbrite (CI) volcanic eruption. We analyzed the genomes of ~35,000-year-old human remains from Crimea and characterized the genetic affiliations with the rare genomes available around the time of the CI eruption to propose the populational history of the peopling of Europe during the Upper Paleolithic (Bennett et al., 2023). 2) The colonization around 7,000 years ago of western Europe by early Neolithic farmers of Anatolian descent who admixed with the autochthonous Mesolithic hunter-gatherers and introduced the producing lifestyle of farming societies that is the basis of our present-day societies. We started this research line with a first analysis of low coverage genomes covering several areas of southern, eastern and northern France and 7,000 years (Brunel et al., 2020). We then produced around 100 genomes from Neolithic archaeological sites in the Paris Basin and reconstructed the evolution of their corresponding populations, along with the gene flow, admixture and migrations that took place during this period. This study lends us broader insight into the evolution of the corresponding populations and societies since the Neolithic represents a key period for the development of societies leading to the present. (Parasayan et al., in prep.) 3) The introduction into western Europe 4,500 years ago of the ancestry of the European steppes represents the ultimate shaping event of the European genome. The population genetic analysis of the genomes of a collective burial in the Paris Basin and the reconstruction of the genome of a missing ancestor whose steppe-ancestry was introduced into a Neolithic community allowed us to show the admixture dynamics between descendants of the nomads of the steppes and Neolithic farmers during this major cultural transition (Parasayan et al., 2024). 4) Finally, we produced the genomes of a medieval population from western France prior and after a period of famine, war and epidemics in the 14th c. CE (Martin et al., in prep; Parasayan et al., in prep).
3. We invested a great deal into the development of methods applied to ancient DNA research.
We invented key adaptations of library construction methods that allowed us to construct very efficiently in a high throughput format several thousands of genomic libraries from highly degraded samples while avoiding most of the material losses, inevitable when using current methods. We applied some of our methodological developments, in the framework of a collaboration with a biomedical research team, to detect circulating DNA in blood, a non-invasive approach used to monitor tumor progression. This contributed to the discovery of cell-free mitochondria circulating in human blood.
Our research projects are in direct continuity of our achievements and are motivated by the questions about recent genome evolution of domestic animals and humans since the Upper Paleolithic, discussed in the previous section. Our novel library construction method adapted to traces of degraded DNA, to be patented and published in the coming years, allowed us to obtain several hundreds of complete ancient genomes up to 120,000 years old. We will now focus on the study of the evolution of these genomes, in particular on the identification of the genomic regions under selection in the last 10,000 years using genomic time series to tackle the following questions:
1. Genomic analyses of the evolution of aurochs towards domesticated cattle. The aurochs, the wild ancestor of domesticated cattle, went extinct in the 17th century in Central Europe, and largely before in most of the uninhabited areas at the early Holocene. Its domestication was initiated in south-west Asia in a region encompassing present-day Turkey, Syria and Iraq. We have produced 72 high coverage ancient aurochs genomes dated between 50,000 to 4,500 years ago and from domesticated cattle between 8,500 years ago to the Middle ages and covering south-west, west and south-east Europe, Anatolia, the Caucasus and the Nile valley. They represent the various domestication stages through the original wild animals, early domesticates and later ones and retrace thus the evolution toward specific ancient and modern breeds. To document better diversity of present-day cattle breeds that were not intensively selected over the last 200 years, we also produced five modern hardy breeds genomes to complement the several hundreds of modern-day cattle genomes that constitute our dataset to follow the genomic changes occurring as a response to cattle domestication. These genomes are now being placed in the extended evolutionary population framework that we have constructed based on mitogenomes. We will analyze in depth our genomic time-series dataset to identify the regions under selection at the various stages of the domestication process. We will take into account the diversity of the various aurochs populations that could have participated in the formation of the early domesticate genomes, both in south-west Asia and during the Neolithic migrations from the Aegean regions to western Europe. These events have shaped the gene pool of both humans and domestic animals in prehistoric times. These ancient genomes will be compared to more than 500 modern domestic cattle genomes already available and that we processed in parallel to our ancient genomes. We will use a large diversity of population genomic methods to disentangle population drift, admixture and selection and therefore identify genomic regions under selective pressure. In doing so, we will characterize in time and space the selection exerted by different ancient societies on particular genomic regions shaping the genomic make-up of present-day cattle breeds.
2. Genomic analyses of European bison evolution. In parallel to our analyses of ancient aurochs, we will also explore the European bison, a closely related species, because it was not domesticated, is not yet extinct, and will allow us to contrast the trajectories of the evolutionary changes in response to domestication or to the human exerted pressure on the habitat. To this end, we have generated 23 high coverage ancient European bison genomes from 120,000 years ago to the 17th century. They will be compared to a dataset of 22 present-day European bison and 39 American bison genomes. This will allow us to monitor the genomic changes through time of a wild population that experienced severe bottlenecks due to climate changes and human-exerted pressure. This will be a useful contrast to the understanding and proper interpretations of the genomic changes of the aurochs and domesticated cattle species.
3. Cat genome evolution in response to domestication. The second domestication model we will study thoroughly at the genomic scale is the cat. Intensive trait selection occurred in this species mostly since the 19th century only, even though certain behavioral traits were likely selected early on but not necessarily due to human intention. The underlying genomic events associated with the history of cats interacting with humans will be characterized through the analysis of 99 generated high-coverage ancient genomes spanning the last 10,000 years, as well as 9 modern day genomes from wild cats from both continental France and Corsica. We will compare our data set to a dataset of 375 modern cat genomes that we have processed in parallel using similar population genomic methods as for the cattle genome project.
4. Paleogenomics of the evolution of disease susceptibility and resistance on the territory of present-day France since the Neolithic. We will study the evolution of the human genome in the same period, in response to the lifestyle changes that followed the shift from hunter-gatherer to farmers, i.e., gene-culture co-evolution associated with lifestyle changes (starch-rich and less diverse diet, famine, immunity response to infectious diseases). The ultimate goal is to understand how human cultural evolution may have driven human biological evolution. Indeed, this lifestyle changes affected the diet and the exposure to pathogens, both following the increased human population density, the increase in long-distance exchanges through time, and the interaction with animals, in particular due to domestication, but also in response to the expansion of the human habitats encroaching into that of wild animals. We will use both the more than a hundred high-coverage Neolithic and medieval human genomes from France we have produced, but we will also include a number of additional genomes produced in the human paleogenomics field. The number of human genomes available is largely superior to that available for domesticated species, and thus the power of the selection detection methods is higher. Even though the human genomic field is much more active and competitive, we still expect to contribute significantly thanks to both the genomic time series we have produced enabling us to directly study past selection by tracking allele frequency changes over time. Such data provide information about timing and location of selection that cannot be obtained through analysis of present-day populations. We will perform parallel analyses of the genomes of two domesticated species, cattle and cats, that have evolved concomitantly during the same period and that may have shared with humans some of their pathogens. Analyzing in parallel human and particularly cattle genomes will allow us to retrace the co-evolution of humans and cattle. We will particularly explore the influence of epidemics, including bovine zoonoses, on the genomes of the populations in the north and west of France from the Neolithic, 7,200 years ago, to the present including the transitions from both hunting-gathering to farming and from small communities to state-level-societies.
Team leaders
 Eva-Maria GEIGL, Emeritus researcher, GRANGE/GEIGL LAB+33 (0)1 57 27 81 32, room 583B
Eva-Maria GEIGL, Emeritus researcher, GRANGE/GEIGL LAB+33 (0)1 57 27 81 32, room 583B Thierry GRANGE, Emeritus researcher, GRANGE/GEIGL LAB+33 (0)1 57 27 81 29, room 583B
Thierry GRANGE, Emeritus researcher, GRANGE/GEIGL LAB+33 (0)1 57 27 81 29, room 583B
To contact a member of the team by e-mail: name.surname@ijm.fr
Major Publications of the team
Parasayan, O., Laurelut, C., Bole, C., Bonnabel, L., Corona, A., Domenech-Jaulneau, C., Paresys, C., Richard, I., Grange, T.*, Geigl, E.-M.* (2024) Late Neolithic collective burial reveals admixture dynamics during the third millennium BCE and the shaping of the European genome Sci. Adv. 10, eadl2468 (2024). Doi: 10.1126/sciadv.adl2468
Bennett, E.A., Parasayan, O., Prat, S., Péan, S., Crépin, L., Yanevich, A., Grange, T.,*, Eva-Maria Geigl, E.-M.* Genome sequences of 36,000 to 37,000 year-old modern humans at Buran-Kaya III in Crimea. (2023) Nature Ecology and Evolution 7, 2160–2172. https://doi.org/10.1038/s41559-023-02211-9
Bennett, E.A., Weber, J., Bendhafer, W., Champlot, S., Peters, J., Schwartz, G., Grange, T., Geigl, E.-M. (2022) The genetic identity of the earliest human-made hybrid animals, the kungas of Syro-Mesopotamia. Science Advances 8, eabm0218. DOI : 10.1126/sciadv.abm0218
Guimaraes, S., Arbuckle, B., Peters, J., Adcock, S., Buitenhuis, H., Chavin, H., Manaseryan, N., Uerpmann, H.-P., Grange, T.*, Geigl, E.-M.* (2020) Ancient DNA shows domestic horses were introduced in the southern Caucasus and Anatolia during the Bronze Age. Science Advances, 6: eabb0030, DOI: 10.1126/sciadv.abb0030
Brunel, S., Bennett, E.A., Cardin, L., Garraud, D., Barrand Emam, H., Beylier, A., Boulestin, B., Chenal, F., Cieselski, E., Convertini, F., Dedet, B., Desenne, S., Dubouloz, J., Duday, H., Fabre, V., Gailledrat, E., Gandelin, M., Gleize, Y., Goepfert, S., Guilaine, J., Hachem, L., Ilett, M., Lambach, F., Mazière, F., Perrin, B., Plouin, S., Pinard, E., Praud, I., Richard, I., Riquier, V., Roure, R., Sendra, B., Thevenet, C., Thiol, S., Vauquelin, E., Vergnaud, L., Grange, T.*, Geigl, E.-M.*, Pruvost, M.* (2020) Ancient genomes from present-day France unveil 7,000 years of its demographic history. Proceedings of the National Academy of Sciences USA, 117(23):12791-12798, DOI:10.1073/pnas.1918034117.
Bennett, E.A., Crevecoeur, I., Viola, B., Derevianko, A.P., Shunkov, M.V., Grange, T., Maureille, B., Geigl, E.-M. (2019) Morphology of the Denisovan phalanx closer to modern humans than to Neandertals. Science Advances, 5:eaaw3950, DOI 10.1126/sciadv.aaw3950, DOI: 10.1126/sciadv.aaw3950
Grange, T., Brugal, J.-P., Flori, L., Gautier, M., Uzunidis, A., and Geigl, E.-M. (2018) The Evolution and Population Diversity of Bison in Pleistocene and Holocene Eurasia: Sex Matters. Diversity 10, 65; doi:10.3390/d10030065
Ottoni, C., Van Neer, W., De Cupere, B., Daligault, J., Guimaraes, S., Peters, J., Spassov, N., Prendergast, M.E., Boivin, N., Morales-Muniz, A., Bălăşescu, A., Becker, C., Benecke, N., Boronenanț, A., Buitenhuis, H., Chahoud, J., Crowther, A., Llorente, L., Manaseryan, N., Monchot, H., Onar, V., Osypińska, M., Putelat, O., Studer, J., Wierer, U., Decorte, R., Grange, T.*, Geigl, E.-M.* (2017) The paleogenetics of cat dispersal in the ancient world. Nature Ecology & Evolution 1, 0139. DOI: https://doi.org/10.1038/s41559-017-0139.
Bennett, E.A., Champlot, S., Peters, J., Arbuckle, B.S., Guimaraes, S., Pruvost, M., Bar-David, S., Davis, S.J.M., Gautier, M., Kaczensky, P., Kuehn, R., Mashkour, M., Morales-Muñiz, M., Pucher, E., Tournepiche, J.-F., Uerpmann, H.-P., Bălăşescu, A., Germonpré, M., Y. Günden, C., Hemami, M.-R., Moullé, P.-E., Öztan, A., Uerpmann, M., Walzer, C., Grange, T.*, Geigl, E.-M.* (2017) Taming the Late Quaternary phylogeography of the Eurasiatic wild ass through ancient and modern DNA. Plos one 12(4): e0174216. /doi.org/10.1371/journal.pone.0174216
Guimaraes S, Pruvost M, Daligault J, Stoetzel E, Bennett EA, Côté NM, Nicolas V, Lalis A, Denys C, Geigl E.-M.*, Grange T.* (2017) A cost-effective high-throughput metabarcoding approach powerful enough to genotype ~44 000 year-old rodent remains from Northern Africa. Mol Ecol Resour. 17: 405-417. doi: 10.1111/1755-0998.12565
Massilani, D., Guimaraes, S., Brugal, J.-P., Bennett, E.A., Tokarska, M., Arbogast, R., Baryshnikov, G., Boeskorov, G., Castel, J.-C., Davydov, S., Madelaine, S., Putelat, O., Spasskaya, N., Uerpmann, H.-P., Grange, T.*, Geigl,E.-M.* (2016) Past climate changes, population dynamics and the origin of Bison in Europe. BMC Biology 14:93-110. DOI 10.1186/s12915-016-0317-7
Côté, N.M.L., Daligault, J., Pruvost, M., Bennett, E.A., Gorgé, O., Guimaraes, S., Capelli, N., Le Bailly, M., Geigl, E.-M.*, Grange, T.* (2016) A New High-Througput Approach to Genotype Ancient Human Gastrointestinal Parasites. PLoS One. 2016 11(1):e0146230. doi: 10.1371/journal.pone.0146230. eCollection 2016.
Bennett, E.A., Massilani, D., Lizzo, G., Daligault, J., Geigl, E.-M., Grange, T. (2014) Library construction for ancient genomics: single strand or double strand? Biotechniques 56:289-300. DOI: 10.2144/000114176
Grange T. and Lourenço E. E. (2011) Mechanisms of Epigenetic Gene Activation in Disease: Dynamics of DNA Methylation and Demethylation, in Epigenetic Aspects of Chronic Diseases; pp 55-73; Roach H.I., Bronner F. and Oreffo R. O. eds, Springer, London.
Charruau P, Fernandes C, Orozco-Terwengel P, Peters J, Hunter L, Ziaie H, Jourabchian A, Jowkar H, Schaller G, Ostrowski S, Vercammen P, Grange T, Schlötterer C, Kotze A, Geigl, E.-M., Walzer C, Burger PA. (2010) Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: evidence for long-term geographic isolates. Mol Ecol. (4):706-724
Champlot S., Berthelot C., Pruvost M., Bennett E. A., Grange T. and Geigl E.M. (2010). An Efficient Multistrategy DNA Decontamination Procedure of PCR reagents for Hypersensitive PCR Applications. PLOS One, 5(9): e13042.
Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S. (2009) Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 29, 150-156
Grange T. (2008) Sensitive Detection of mRNA Decay Products Using Reverse-Ligation-Mediated PCR (RL-PCR). in Methods in Enzymology, RNA Turnover in Eukaryotes, Part B, 448, 445-466
Pruvost, M., Schwarz, R., Bessa Correia, V., Champlot, S., Grange, T. and Geigl, E.-M. (2008). DNA diagenesis and palaeogenetic analysis: critical assessment and methodological progress. Palaeogeography, Palaeoclimatology, Palaeoecology, 266, 211-219.
Miele V., C. Vaillant, Y. d’Aubenton-Carafa, C. Thermes and T. Grange (2008). DNA physical properties determine nucleosome occupancy from yeast to fly. Nucl Acids Res, 36, 3746-3756
Pruvost, M., T. Grange and E. M. Geigl (2005). “Minimizing DNA contamination by using UNG-coupled quantitative real-time PCR on degraded DNA samples: application to ancient DNA studies.” Biotechniques 38(4): 569-75.
abstract
Thomassin, H., C. Kress and T. Grange (2004). “MethylQuant: a sensitive method for quantifying methylation of specific cytosines within the genome.” Nucleic Acids Res 32(21).
abstract
Flavin, M., L. Cappabianca, C. Kress, H. Thomassin and T. Grange (2004). “Nature of the accessible chromatin at a glucocorticoid-responsive enhancer.” Mol Cell Biol 24(18): 7891-901.
abstract
Pruvost, M. and E. M. Geigl (2004). “Real-time quantitative PCR to assess the authenticity of ancient DNA amplification.” J. Archaeol. Sci. 31(9): 1191-1197.
abstract
Couttet, P. and T. Grange (2004). “Premature termination codons enhance mRNA decapping in human cells.” Nucleic Acids Res 32(2): 488-94. Print 2004.
abstract
Grange, T., L. Cappabianca, M. Flavin, H. Sassi and H. Thomassin (2001). “In vivo analysis of the model tyrosine aminotransferase gene reveals multiple sequential steps in glucocorticoid receptor action.” Oncogene 20(24): 3028-38.
abstract
Geigl, E.-M. (2002) “On the Circumstances Surrounding the Preservation and Analysis of Very Old DNA”. Archaeometry 44 (3) :337-342,
Thomassin, H., M. Flavin, M. L. Espinas and T. Grange (2001). “Glucocorticoid-induced DNA demethylation and gene memory during development.” Embo J 20(8): 1974-83.
abstract
Geigl, E. M. (2001). “Inadequate use of molecular hybridization to analyze DNA in Neanderthal fossils.” Am J Hum Genet 68(1): 287-91.
abstract
Christoffels, V. M., H. Sassi, J. M. Ruijter, A. F. Moorman, T. Grange and W. H. Lamers (1999). “A mechanistic model for the development and maintenance of portocentral gradients in gene expression in the liver.” Hepatology 29(4): 1180-92.
abstract
Sassi, H., R. Pictet and T. Grange (1998). “Glucocorticoids are insufficient for neonatal gene induction in the liver.” Proc Natl Acad Sci U S A 95(10): 5621-5.
abstract
Couttet, P., M. Fromont-Racine, D. Steel, R. Pictet and T. Grange (1997). “Messenger RNA deadenylylation precedes decapping in mammalian cells.” Proc Natl Acad Sci U S A 94(11): 5628-33.
abstract
De Sario, A., Geigl, E.-M., Palmieri, G., D’Urso, M., Bernardi, G.: A compositional map of human chromosome band Xq28. Proc. Natl. Acad. Sci. USA 93:1298-1302, 1996
Francke, U., Chang, E., Comeau, K., Geigl, E.-M., Giacalone, J., Li, X., Luna, J., Moon, A., Welch, S., Wilgenbus, P. (auteurs listés par ordre alphabétique) (1994) A radiation hybrid map of human chromosome 18. Cytogenet Cell Genet 66:196-213.
Leib-Mösch, C., Haltmeier, M., Werner, T., Geigl, E.-M., Brack-Werner, R., Francke, U., Erfle, V., Hehlmann, R. (1993) Genomic distribution and transcription of solitary HERV-K LTRs. Genomics 18:261-269.
Geigl, E.-M., Eckardt-Schupp, F. (1990) Chromosome-specific identification and quantification of S1 nuclease-sensitive sites in yeast chromatin by pulsed field gel electrophoresis. Molecular Microbiology 4(5):801-810.
Publications Thierry Grange
Kaptan D, Atağ G, Vural KB, Morell Miranda P, Akbaba A, Yüncü E, Buluktaev A, Abazari MF, Yorulmaz S, Kazancı DD, Küçükakdağ Doğu A, Çakan YG, Özbal R, Gerritsen F, De Cupere B, Duru R, Umurtak G, Arbuckle BS, Baird D, Çevik Ö, Bıçakçı E, Gündem CY, Pişkin E, Hachem L, Canpolat K, Fakhari Z, Ochir-Goryaeva M, Kukanova V, Valipour HR, Hoseinzadeh J, Küçük Baloğlu F, Götherström A, Hadjisterkotis E, Grange T, Geigl EM, Togan İZ, Günther T, Somel M, Özer F. The Population History of Domestic Sheep Revealed by Paleogenomes. Mol Biol Evol. 2024 Oct 4;41(10):msae158. doi: 10.1093/molbev/msae158. PMID: 39437846; PMCID: PMC11495565.
Latourte A, Jaulerry S, Combier A, Cherifi C, Jouan Y, Grange T, Daligault J, Ea HK, Cohen-Solal M, Hay E, Richette P. Ann Rheum Dis. (2024) SerpinA3N limits cartilage destruction in osteoarthritis by inhibiting macrophage-derived leucocyte elastase. ard-2024-225645. doi: 10.1136/ard-2024-225645. Online ahead of print. PMID: 39134394
Özkan M, Gürün K, Yüncü E, Vural KB, Atağ G, Akbaba A, Fidan FR, Sağlıcan E, Altınışık EN, Koptekin D, Pawłowska K, Hodder I, Adcock SE, Arbuckle BS, Steadman SR, McMahon G, Erdal YS, Bilgin CC, Togan İ, Geigl EM, Götherström A, Grange T, Özer F, Somel M. (2024) The first complete genome of the extinct European wild ass (Equus hemionus hydruntinus). Mol Ecol. 2024 Jul 1:e17440. doi: 10.1111/mec.17440.
Parasayan, O., Laurelut, C., Bole, C., Bonnabel, L., Corona, A., Domenech-Jaulneau, C., Paresys, C., Richard, I., Grange, T.*, Geigl, E.-M.* (2024) Late Neolithic collective burial reveals admixture dynamics during the third millennium BCE and the shaping of the European genome Sci. Adv. 10, eadl2468 (2024). Doi: 10.1126/sciadv.adl2468
Andrew Bennett, Oğuzhan Parasayan, Sandrine Prat, Stéphane Péan, Laurent Crépin, Alexandr Yanevich, Thierry Grange*, Eva-Maria Geigl* Genome sequences of 36,000 to 37,000 year-old modern humans at Buran-Kaya III in Crimea. (2023) Nature Ecology and Evolution 7, 2160–2172. https://doi.org/10.1038/s41559-023-02211-9
Jamieson A, Carmagnini A, Howard-McCombe J, Doherty S, Hirons A, Dimopoulos E, Lin AT, Allen R, Anderson-Whymark H, Barnett R, Batey C, Beglane F, Bowden W, Bratten J, De Cupere B, Drew E, Foley NM, Fowler T, Fox A, Geigl EM, Gotfredsen AB, Grange T, Griffiths D, Groß D, Haruda A, Hjermind J, Knapp Z, Lebrasseur O, Librado P, Lyons LA, Mainland I, McDonnell C, Muñoz-Fuentes V, Nowak C, O’Connor T, Peters J, Russo IM, Ryan H, Sheridan A, Sinding MS, Skoglund P, Swali P, Symmons R, Thomas G, Trolle Jensen TZ, Kitchener AC, Senn H, Lawson D, Driscoll C, Murphy WJ, Beaumont M, Ottoni C, Sykes N, Larson G, Frantz L. (2023) Limited historical admixture between European wildcats and domestic cats. Curr Biol. 33(21):4751-4760.e14. doi: 10.1016/j.cub.2023.08.031
Gorgé O., Bennett EA., Daligault J., Massilani D., Geigl E.-M.*, Grange T.* (2022) Analysis of ancient microbial DNA. Microbial Environmental Genomic, Methods Mol Biol 2605:103-131, doi: 10.1007/978-1-0716-2871-3_6
Bennett, E.A., Weber, J., Bendhafer, W., Champlot, S., Peters, J., Schwartz, G., Grange, T., Geigl, E.-M. (2022) The genetic identity of the earliest human-made hybrid animals, the kungas of Syro-Mesopotamia. Science Advances 8, eabm0218. DOI : 10.1126/sciadv.abm0218
Guimaraes, S., Arbuckle, B., Peters, J., Adcock, S., Buitenhuis, H., Chavin, H., Manaseryan, N., Uerpmann, H.-P., Grange, T.*, Geigl, E.-M.* (2020) Ancient DNA shows domestic horses were introduced in the southern Caucasus and Anatolia during the Bronze Age. Science Advances, 6: eabb0030, DOI: 10.1126/sciadv.abb0030
Al Amir Dache Z., Otandault A., Tanos R., Pastor B., Meddeb R., Sanchez C., Arena G., Lasorsa L., Bennett E.A., Grange T., El Messaoudi S., Mazard T., Prevostel C., Thierry A.R. (2020) Blood contains circulating cell-free respiratory competent mitochondria. FASEB J;34(3):3616-3630. doi: 10.1096/fj.201901917RR. Epub 2020 Jan 19.
Brunel, S., Bennett, E.A., Cardin, L., Garraud, D., Barrand Emam, H., Beylier, A., Boulestin, B., Chenal, F., Cieselski, E., Convertini, F., Dedet, B., Desenne, S., Dubouloz, J., Duday, H., Fabre, V., Gailledrat, E., Gandelin, M., Gleize, Y., Goepfert, S., Guilaine, J., Hachem, L., Ilett, M., Lambach, F., Mazière, F., Perrin, B., Plouin, S., Pinard, E., Praud, I., Richard, I., Riquier, V., Roure, R., Sendra, B., Thevenet, C., Thiol, S., Vauquelin, E., Vergnaud, L., Grange, T.*, Geigl, E.-M.*, Pruvost, M.* (2020) Ancient genomes from present-day France unveil 7,000 years of its demographic history. Proceedings of the National Academy of Sciences USA, 117(23):12791-12798, DOI:10.1073/pnas.1918034117.
Bennett, E.A., Crevecoeur, I., Viola, B., Derevianko, A.P., Shunkov, M.V., Grange, T., Maureille, B., Geigl, E.-M. (2019) Morphology of the Denisovan phalanx closer to modern humans than to Neandertals. Science Advances, 5:eaaw3950, DOI 10.1126/sciadv.aaw3950, DOI: 10.1126/sciadv.aaw3950
Geigl, E.-M., Grange, T. (2018) Ancient DNA: The quest for the best. Mol Ecol Resour. 18:1185–1187. DOI: 10.1111/1755-0998.12931
Grange, T., Brugal, J.-P., Flori, L., Gautier, M., Uzunidis, A., and Geigl, E.-M. (2018) The Evolution and Population Diversity of Bison in Pleistocene and Holocene Eurasia: Sex Matters. Diversity 10, 65; doi:10.3390/d10030065
Ottoni, C., Van Neer, W., De Cupere, B., Daligault, J., Guimaraes, S., Peters, J., Spassov, N., Prendergast, M.E., Boivin, N., Morales-Muniz, A., Bălăşescu, A., Becker, C., Benecke, N., Boronenanț, A., Buitenhuis, H., Chahoud, J., Crowther, A., Llorente, L., Manaseryan, N., Monchot, H., Onar, V., Osypińska, M., Putelat, O., Studer, J., Wierer, U., Decorte, R., Grange, T.*, Geigl, E.-M.* (2017) The paleogenetics of cat dispersal in the ancient world. Nature Ecology & Evolution 1, 0139. DOI: https://doi.org/10.1038/s41559-017-0139.
Bennett, E.A., Champlot, S., Peters, J., Arbuckle, B.S., Guimaraes, S., Pruvost, M., Bar-David, S., Davis, S.J.M., Gautier, M., Kaczensky, P., Kuehn, R., Mashkour, M., Morales-Muñiz, M., Pucher, E., Tournepiche, J.-F., Uerpmann, H.-P., Bălăşescu, A., Germonpré, M., Y. Günden, C., Hemami, M.-R., Moullé, P.-E., Öztan, A., Uerpmann, M., Walzer, C., Grange, T.*, Geigl, E.-M.* (2017) Taming the Late Quaternary phylogeography of the Eurasiatic wild ass through ancient and modern DNA. Plos one 12(4): e0174216. /doi.org/10.1371/journal.pone.0174216
Guimaraes S, Pruvost M, Daligault J, Stoetzel E, Bennett EA, Côté NM, Nicolas V, Lalis A, Denys C, Geigl EM*, Grange T.* (2017) A cost-effective high-throughput metabarcoding approach powerful enough to genotype ~44 000 year-old rodent remains from Northern Africa. Mol Ecol Resour. 17: 405-417. doi: 10.1111/1755-0998.12565
Massilani, D., Guimaraes, S., Brugal, J.-P., Bennett, E.A., Tokarska, M., Arbogast, R., Baryshnikov, G., Boeskorov, G., Castel, J.-C., Davydov, S., Madelaine, S., Putelat, O., Spasskaya, N., Uerpmann, H.-P., Grange, T.*, Geigl,E.-M.* (2016) Past climate changes, population dynamics and the origin of Bison in Europe. BMC Biology 14:93-110. DOI 10.1186/s12915-016-0317-7
Gorgé O, Bennett EA, Massilani D, Daligault J, Pruvost M, Geigl E.-M., Grange T. (2016) Analysis of Ancient DNA in Microbial Ecology. Methods Mol Biol. 1399:289-315. doi: 10.1007/978-1-4939-3369-3_17.
Côté, N.M.L., Daligault, J., Pruvost, M., Bennett, E.A., Gorgé, O., Guimaraes, S., Capelli, N., Le Bailly, M., Geigl*, E.-M., Grange, T.* (2016) A New High-Througput Approach to Genotype Ancient Human Gastrointestinal Parasites. PLoS One. 2016 11(1):e0146230. doi: 10.1371/journal.pone.0146230. eCollection 2016.
Guimaraes, S., Fernandez-Jalvo, Y., Stoetzel, E., Gorgé, O., Bennett, E.A., Denys, C., Grange, T., Geigl, E.-M. (2016) Owl pellets: a wise DNA source for small mammal genetics. Journal of Zoology 298(1):64–74. doi:10.1111/jzo.12285
Bennett, E.A., Massilani, D., Lizzo, G., Daligault, J., Geigl, E.-M., Grange, T. (2014) Library construction for ancient genomics: single strand or double strand? Biotechniques 56:289-300. DOI: 10.2144/000114176
Geigl E. M. and Grange T. (2012) Eurasian wild asses in time and space: Morphological versus genetic diversity. Ann Anat. 194:88-102
Geigl, E.M., Grange, T., Maureille, B. : Le génome néandertalien. Encyclopaedia Universalis, « La Science au présent 2011 », p. 133-140
Charruau P, Fernandes C, Orozco-Terwengel P, Peters J, Hunter L, Ziaie H, Jourabchian A, Jowkar H, Schaller G, Ostrowski S, Vercammen P, Grange T, Schlötterer C, Kotze A, Geigl EM, Walzer C, Burger PA. (2011) Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: evidence for long-term geographic isolates. Mol Ecol. (4):706-724
Grange T. and Lourenço E. (2011) Mechanisms of Epigenetic Gene Activation in Disease: Dynamics of DNA Methylation and Demethylation, in Epigenetic Aspects of Chronic Diseases; pp 55-73; Roach H.I., Bronner F. and Oreffo R. O. eds, Springer, London.
Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S. (2009) Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 29, 150-156
Champlot S., Berthelot C., Pruvost M., Bennett E. A., Grange T. and Geigl E.M. (2010). An Efficient Multistrategy DNA Decontamination Procedure of PCR reagents for Hypersensitive PCR Applications. PLOS One, 5(9): e13042.
Grange T. (2008) Sensitive Detection of mRNA Decay Products Using Reverse-Ligation-Mediated PCR (RL-PCR). In Methods in Enzymology, RNA Turnover in Eukaryotes, Part B, 448, 445-466
Pruvost, M., Schwarz, R., Bessa Correia, V., Champlot, S., Grange, T. and Geigl, E.-M.(2008) DNA diagenesis and palaeogenetic analysis: critical assessment and methodological progress. Palaeogeography, Palaeoclimatology, Palaeoecology, 266, 211-219.
Miele V., C. Vaillant, Y. d’Aubenton-Carafa, C. Thermes and T. Grange (2008) DNA physical properties determine nucleosome occupancy from yeast to fly. Nucl. Acids Res, 36, 3746-3756
Dugast-Darzacq C., and T. Grange (2008) MethylQuant : a real time PCR based method to quantify methylation at single specific cytosines. In Methods in Molecular Biology, DNA methylation: Methods and protocols, Second edition, J. Tost Ed, Humana Press, 507, 281-303.
Dugast-Darzacq C., T. Grange and N. B. Schreiber-Agus (2007) Differential effects of Mxi1-SR and Mxi1-SR in Myc antagonism. FEBS J. 274, 4643-53.
Pruvost, R. Schwarz, V. Bessa Correia, S. Champlot, S. Braguier, N. Morel, Y. Fernandez-Jalvo, T. Grange and EM Geigl (2007) Freshly excavated fossil bones are best for amplification of ancient DNA M. Proc. Ntl. Acad. Sci. USA, 104, 739-744
Hoogenkamp M, Gaemers IC, Schoneveld OJ, Das AT, Grange T, Lamers WH (2007) Hepatocyte-specific interplay of transcription factors at the far-upstream enhancer of the carbamoylphosphate synthetase gene upon glucocorticoid induction. FEBS J. 274, 37–45
Schalchi L., Grange T., Hänni, C., and Morange M. (2006) Bac to basics: l’ADN. La Recherche, 398, 75-78
Active cytosine demethylation triggered by a transcriptional regulator involves DNA strand breaks. Kress C., Thomassin H., and Grange T., (2006) Proc. Ntl. Acad. Sci. USA 103, 1112-1117.
Grange, T., J. Imbert and D. Thieffry (2005). “Epigenomics: large scale analysis of chromatin modifications and transcription factors/genome interactions.” Bioessays 27(11): 1203-5.
abstract
Flavin, M., L. Cappabianca, C. Kress, H. Thomassin and T. Grange (2004). “Nature of the accessible chromatin at a glucocorticoid-responsive enhancer.” Mol Cell Biol 24(18): 7891-901.
abstract
Thakur, N., V. K. Tiwari, H. Thomassin, R. R. Pandey, M. Kanduri, A. Gondor, T. Grange, R. Ohlsson and C. Kanduri (2004). “An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region.” Mol Cell Biol 24(18): 7855-62.
abstract
Thomassin, H., C. Kress and T. Grange (2004). “MethylQuant: a sensitive method for quantifying methylation of specific cytosines within the genome.” Nucleic Acids Res 32(21).
abstract
Pâques F. and T. Grange (2002). Architecture nucléaire et régulation transcriptionnelle chez les eucaryotes supérieurs. Med./Sci 18, 1245-1256.
abstract
Grange, T. (2001). Traduction nucléaire : un garde-fou contre les protéines tronquées ?
Med./Sci. 17, 914-915.
abstract
Grange, T., L. Cappabianca, M. Flavin, H. Sassi and H. Thomassin (2001). “In vivo analysis of the model tyrosine aminotransferase gene reveals multiple sequential steps in glucocorticoid receptor action.” Oncogene 20(24): 3028-38.
abstract
Kress, C., H. Thomassin and T. Grange (2001). “Local DNA demethylation in vertebrates: how could it be performed and targeted'” FEBS Lett 494(3): 135-40.
abstract
Thomassin, H., M. Flavin, M. L. Espinas and T. Grange (2001). “Glucocorticoid-induced DNA demethylation and gene memory during development.” Embo J 20(8): 1974-83.
abstract
Cappabianca, L., H. Thomassin, R. Pictet and T. Grange (1999). “Genomic footprinting using nucleases.” Methods Mol Biol 119: 427-42.
abstract
Christoffels, V. M., H. Sassi, J. M. Ruijter, A. F. Moorman, T. Grange and W. H. Lamers (1999). “A mechanistic model for the development and maintenance of portocentral gradients in gene expression in the liver.” Hepatology 29(4): 1180-92.
abstract
Grange T. and H. Thomassin (1999). « Comment déméthyler une cytosine? Enlève-t-on un peu, beaucoup, encore plus, ou pas du tout’ » Med./Sci., 15, 907-908.
abstract
Thomassin, H., E. J. Oakeley and T. Grange (1999). “Identification of 5-methylcytosine in complex genomes.” Methods 19(3): 465-75.
abstract
Christoffels, V. M., T. Grange, K. H. Kaestner, T. J. Cole, G. J. Darlington, C. M. Croniger and W. H. Lamers (1998). “Glucocorticoid receptor, C/EBP, HNF3, and protein kinase A coordinately activate the glucocorticoid response unit of the carbamoylphosphate synthetase I gene.” Mol Cell Biol 18(11): 6305-15.
abstract
Sassi, H., R. Pictet and T. Grange (1998). “Glucocorticoids are insufficient for neonatal gene induction in the liver.” Proc Natl Acad Sci U S A 95(10): 5621-5.
abstract
Bertrand, E., D. Castanotto, C. Zhou, C. Carbonnelle, N. S. Lee, P. Good, S. Chatterjee, T. Grange, R. Pictet, D. Kohn, D. Engelke and J. J. Rossi (1997). “The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization.” Rna 3(1): 75-88.
abstract
Bertrand, E., M. Fromont-Racine, R. Pictet and T. Grange (1997). “Detection of ribozyme cleavage products using reverse ligation-mediated PCR (RL-PCR).” Methods Mol Biol 74: 311-23.
abstract
Couttet, P., M. Fromont-Racine, D. Steel, R. Pictet and T. Grange (1997). “Messenger RNA deadenylylation precedes decapping in mammalian cells.” Proc Natl Acad Sci U S A 94(11): 5628-33.
abstract
Grange T., G. Rigaud, E. Bertrand, M. Fromont-Racine, M.-L. Espinás, J. Roux and R. Pictet (1997). “In vivo footprinting of the interaction of proteins with DNA and RNA.” in In vivo footprinting. Cartwright, I. L. Ed. Advances in Molecular and Cell Biology. JAI Press, Inc., Greenwich, Conn. 21, 73-109.
Grange, T., E. Bertrand, M. L. Espinas, M. Fromont-Racine, G. Rigaud, J. Roux and R. Pictet (1997). “In vivo footprinting of the interaction of proteins with DNA and RNA.” Methods 11(2): 151-63.
abstract
Espinas, M. L., J. Roux, R. Pictet and T. Grange (1995). “Glucocorticoids and protein kinase A coordinately modulate transcription factor recruitment at a glucocorticoid-responsive unit.” Mol Cell Biol 15(10): 5346-54.
abstract
Roux, J., R. Pictet and T. Grange (1995). “Hepatocyte nuclear factor 3 determines the amplitude of the glucocorticoid response of the rat tyrosine aminotransferase gene.” DNA Cell Biol 14(5): 385-96.
abstract
Sassi, H., M. Fromont-Racine, T. Grange and R. Pictet (1995). “Tissue specificity of a glucocorticoid-dependent enhancer in transgenic mice.” Proc Natl Acad Sci U S A 92(16): 7197-201.
abstract
Bertrand, E., M. Fromont-Racine, R. Pictet and T. Grange (1993). “Visualization of the interaction of a regulatory protein with RNA in vivo.” Proc Natl Acad Sci U S A 90(8): 3496-500.
abstract
Bertrand, E., R. Pictet and T. Grange (1994). “Can hammerhead ribozymes be efficient tools to inactivate gene function?” Nucleic Acids Res 22(3): 293-300.
abstract
Espinas, M. L., J. Roux, J. Ghysdael, R. Pictet and T. Grange (1994). “Participation of Ets transcription factors in the glucocorticoid response of the rat tyrosine aminotransferase gene.” Mol Cell Biol 14(6): 4116-25.
abstract
Schweizer-Groyer, G., A. Groyer, F. Cadepond, T. Grange, E. E. Baulieu and R. Pictet (1994). “Expression from the tyrosine aminotransferase promoter (nt -350 to 1) is liver-specific and dependent on the binding of both liver-enriched and ubiquitous trans-acting factors.” Nucleic Acids Res 22(9): 1583-92.
abstract
Fromont-Racine, M., E. Bertrand, R. Pictet and T. Grange (1993). “A highly sensitive method for mapping the 5′ termini of mRNAs.” Nucleic Acids Res 21(7): 1683-4.
abstract
Bertrand E., T. Grange and R. Pictet (1992). “Trans-acting hammerhead ribozymes in vivo: present limits and future directions.” in Gene regulation by antisense RNA and DNA. pp 71-81. R.P. Erickson & J. Izart Eds. Keistone Symposia series on Molecular and Cellular Biology. Raven Press, Ltd., New York.
Schweizer-Groyer, G., A. Groyer, F. Cadepond, T. Grange, E. E. Baulieu and R. Pictet (1992). “Two liver-enriched trans-acting factors support the tissue-specific basal transcription from the rat tyrosine aminotransferase promoter.” J Steroid Biochem Mol Biol 41(3-8): 747-52.
abstract
Grange, T., J. Roux, G. Rigaud and R. Pictet (1991). “Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor.” Nucleic Acids Res 19(1): 131-9.
abstract
Rigaud, G., T. Grange and R. Pictet (1991). “Sequenase should be used instead of the Klenow fragment for the synthesis of oligonucleotides labeled to a high specific activity.” Nucleic Acids Res 19(17): 4777.
abstract
Rigaud, G., J. Roux, R. Pictet and T. Grange (1991). “In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor.” Cell 67(5): 977-86.
abstract
Grange, T., J. Roux, M. Fromont-Racine and R. Pictet (1989). “Positive and negative regulation of a transfected chimeric tyrosine aminotransferase gene: effect of copy number.” Exp Cell Res 180(1): 220-33.
abstract
Grange, T., J. Roux, G. Rigaud and R. Pictet (1989). “Two remote glucocorticoid responsive units interact cooperatively to promote glucocorticoid induction of rat tyrosine aminotransferase gene expression.” Nucleic Acids Res 17(21): 8695-709.
abstract
Oddos, J., T. Grange, K. D. Carr, B. Matthews, J. Roux, H. Richard-Foy and R. Pictet (1989). “Nucleotide sequence of 10 kilobases of rat tyrosine aminotransferase gene 5′ flanking region.” Nucleic Acids Res 17(21): 8877-8.
abstract
Grange, T., C. M. de Sa, J. Oddos and R. Pictet (1987). “Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif.” Nucleic Acids Res 15(12): 4771-87.
abstract
Rigaud, G., T. Grange and R. Pictet (1987). “The use of NaOH as transfer solution of DNA onto nylon membrane decreases the hybridization efficiency.” Nucleic Acids Res 15(2): 857.
abstract
Grange, T., M. Bouloy and M. Girard (1985). “Stable secondary structures at the 3′-end of the genome of yellow fever virus (17 D vaccine strain).” FEBS Lett 188(1): 159-63.
abstract
Grange, T., C. Guenet, J. B. Dietrich, S. Chasserot, M. Fromont, N. Befort, J. Jami, G. Beck and R. Pictet (1985). “Complete complementary DNA of rat tyrosine aminotransferase messenger RNA. Deduction of the primary structure of the enzyme.” J Mol Biol 184(2): 347-50.
abstract
Grange, T., F. Kunst, J. Thillet, B. Ribadeau-Dumas, S. Mousseron, A. Hung, J. Jami and R. Pictet (1984). “Expression of the mouse dihydrofolate reductase cDNA in B. subtilis: a system to select mutant cDNAs coding for methotrexate resistant enzymes.” Nucleic Acids Res 12(8): 3585-601.
abstract
Publications Eva-Maria Geigl
Leib-Mösch, C., Barton, D., Geigl, E.-M., Brack-Werner, R., Erfle, V., Hehlmann, R., Francke, U.: Two RFLPs associated with the human endogenous retroviral element S71 on chromosome 18q21. Nucl. Acid. Res 17 (6):2367, 1988
Hagen, U., Bertram, H., Geigl, E.-M., Kohfeldt, E., Wendel, S.: Radiation-induced clustered damage in DNA. Free Rad. Res. Comm. 6:177-178, 1989
Geigl, E.-M., Eckardt-Schupp, F. (1990) Chromosome-specific identification and quantification of S1 nuclease-sensitive sites in yeast chromatin by pulsed field gel electrophoresis. Molecular Microbiology 4(5):801-810.
Geigl, E.-M., Eckardt-Schupp, F. (1991) The repair of DNA double-strand breaks and S1 nuclease-sensitive sites can be monitored chromosome-specifically in Saccharomyces cerevisiae using pulsed-field gel electrophoresis. Molecular Microbiology, 5(7):1615-1620.
Geigl, E.-M., Eckardt-Schupp, F. (1991) Repair of gamma ray-induced S1 nuclease hypersensitive sites in yeast depends on homologous mitotic recombination and a RAD18-dependent function. Curr. Genet. 20:33-37.
Leib-Mösch, C., Haltmeier, M., Werner, T., Geigl, E.-M., Brack-Werner, R., Francke, U., Erfle, V., Hehlmann, R. (1993) Genomic distribution and transcription of solitary HERV-K LTRs. Genomics 18:261-269,
Francke, U., Chang, E., Comeau, K., Geigl, E.-M., Giacalone, J., Li, X., Luna, J., Moon, A., Welch, S., Wilgenbus, P. (auteurs listés par ordre alphabétique) (1994) A radiation hybrid map of human chromosome 18. Cytogenet Cell Genet 66:196-213,
De Sario, A., Geigl, E.-M., Bernardi, G. (1995) A rapid procedure for the compositional analysis of yeast artificial chromosomes. Nucleic Acids Res. 23(19):4013-4014,
De Sario, A., Geigl, E.-M., Palmieri, G., D’Urso, M., Bernardi, G. (J’ai co-encadré l’étudiante en these A. DeSario): A compositional map of human chromosome band Xq28. Proc. Natl. Acad. Sci. USA 93:1298-1302, 1996
Geigl, E.-M.: “Inadequate Use of Molecular Hybridization to Analyze DNA in Neanderthal Fossils”. Am. J. Hum. Genetics 68:287-290, 2001 ; S0002-9297(07)62499-9/10.1086/316948
Geigl, E.-M. : On the Circumstances Surrounding the Preservation and Analysis of Very Old DNA. Archaeometry 44 (3) :337-342, 2002
Geigl, E.-M., Baumer, U., and Koller, J.: New Appproaches to the study of the preservation of biopolymers in fossil bones. Environ. Chem. Letters 2(1) :45-48, 2004
Pruvost, M. and Geigl, E.-M. : Real-time Quantitative PCR to Assess the Authenticity of Ancient DNA, Journal of Archaeological Science, 31(9):1191-1197, 2004
Geigl, E.-M. and Pruvost, M. : Plea for a multidisciplinary approach to the study of Neolithic migrations : the analysis of biological witnesses and the input of palaeogenetics. In « Colonisation,, Migration, and Marginal Areas. A Zooarchaeological Approach. » M. Mondini, S. Munoz and S. Wickler (eds), Oxbow Books, p. 10-19, 2004.
Geigl, E.-M. : Why ancient DNA research needs taphonomy. In « Biosphere to Lithosphere: new studies in vertebrate taphonomy » T.O’Connor (ed.), Oxbow Books., p. 79-86, 2005.
Geigl, E.-M.: A personal analysis of the high failure rate of ancient DNA research. Geoarchaeological and Bioarchaeological Studies, Volume 3, p. 463-466, 2005.
Pruvost, M., Grange, T., and Geigl, E.-M.: Minimizing DNA-contamination by using UNG-coupled quantitative real-time PCR (UQPCR) on degraded DNA samples: application to ancient DNA studies. Biotechniques 38 :569-575, 2005; DOI 05384ST03
Pruvost, M., Schwarz, R., Bessa Correia, V., Champlot, S., Braguier, S., Morel, N., Fernandez-Jalvo, Y., Grange, T. and Geigl, E.-M.: Freshly excavated fossil bones are best for ancient DNA amplification. Proc. Natl. Acad. Sci. USA. 104 (3): 739-744, 2007 ; DOI 0610257104/ 10.1073/pnas.0610257104
Pruvost, M., Schwarz, R., Bessa Correia, V., Champlot, S., Grange, T. and Geigl, E.-M. DNA diagenesis and palaeogenetic analysis: critical assessment and methodological progress. Palaeogeography, Palaeoclimatology, Palaeoecology, 266 (3-4) : 211-219, 2008. doi:10.1016/j.palaeo.2008.03.041
Geigl, E.-M. : Palaeogenetics of cattle domestication : Methodological challenge for the study of fossil bones preserved in the domestication center in Southwest Asia. Comptes Rendus Palévol 7(2-3): 99-112, 2008
Fernandez-Jalvo, Y., Andrews, P., Pesquero, D., Smith, C., Marin-Monfort, D., Sanchez, B., Geigl, E.-M., Alonso, A. Early bone diagenesis in temperate environments. Part I: Surface features and histology. Palaeogeography, Palaeoclimatology, Palaeoecology 288:62-81, 2010
Champlot, S., Berthelot,C., Pruvost, M., Bennett, E.A., Grange, T., Geigl, E.-M. (2010) An Efficient Multistrategy DNA Decontamination Procedure of PCR reagents for Hypersensitive PCR Applications. PLoS ONE 5(9):e13042,. DOI 10.1371/journal.pone.0013042
Charruau, P., Fernandes, C., Orozco-Ter Wengel, P. Peters, J., Hunter, L., Ziaie, H., Jourabchian, A., Jowkar, H., Schaller, G., Ostrowski, S., Vercammen, P., Grange, T., Schlötterer, C., Kotze, A. Geigl, E.-M., Walzer, C., Burger, P.A. Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: evidence for long-term geographic isolation. Mol. Ecol. 20(4):706-24, 2010. doi: 10.1111/j.1365-294X.2010.04986
Geigl, E.-M. and Grange, T. (2012) Eurasian wild asses in time and space: morphological versus genetic diversity. Annals of Anatomy. 194:88-102,. doi:10.1016/j.aanat.2011.06.002
Bennett, E.A., Massilani, D., Lizzo, G., Daligault, J., Geigl, E.-M., Grange, T. (2014) Library construction for ancient genomics: single strand or double strand? Biotechniques 56:289-300. DOI: 10.2144/000114176
Nores, C., Morales-Muniz, A., Llorente Rodriguez, L., Bennett, E.A., Geigl, E.-M. (2015) The Iberian zebro: what kind of beast was it? Anthropozoologica 50(1):21-32.
Guimaraes, S., Fernandez-Jalvo, Y., Stoetzel, E., Gorgé, O., Bennett, E.A., Denys, C., Grange, T., Geigl, E.-M. (2016) Owl pellets: a wise DNA source for small mammal genetics. Journal of Zoology 298(1):64–74. doi:10.1111/jzo.12285
Côté, N.M.L., Daligault, J., Pruvost, M., Bennett, E.A., Gorgé, O., Guimaraes, S., Capelli, N., Le Bailly, M., Geigl*, E.-M., Grange, T.* (2016) A New High-Througput Approach to Genotype Ancient Human Gastrointestinal Parasites. PLoS One. 2016 11(1):e0146230. doi: 10.1371/journal.pone.0146230. eCollection 2016.
Gorgé O, Bennett EA, Massilani D, Daligault J, Pruvost M, Geigl EM, Grange T. (2016) Analysis of Ancient DNA in Microbial Ecology. Methods Mol Biol. 1399:289-315. doi: 10.1007/978-1-4939-3369-3_17.
Guimaraes S, Pruvost M, Daligault J, Stoetzel E, Bennett EA, Côté NM, Nicolas V, Lalis A, Denys C, Geigl EM*, Grange T.* (2017) A cost-effective high-throughput metabarcoding approach powerful enough to genotype ~44 000 year-old rodent remains from Northern Africa. Mol Ecol Resour. 17: 405-417. doi: 10.1111/1755-0998.12565
Massilani, D., Guimaraes, S., Brugal, J.-P., Bennett, E.A., Tokarska, M., Arbogast, R., Baryshnikov, G., Boeskorov, G., Castel, J.-C., Davydov, S., Madelaine, S., Putelat, O., Spasskaya, N., Uerpmann, H.-P., Grange, T.*, Geigl,E.-M.* (2016) Past climate changes, population dynamics and the origin of Bison in Europe. BMC Biology 14:93-110. DOI 10.1186/s12915-016-0317-7
Bennett, E.A., Champlot, S., Peters, J., Arbuckle, B.S., Guimaraes, S., Pruvost, M., Bar-David, S., Davis, S.J.M., Gautier, M., Kaczensky, P., Kuehn, R., Mashkour, M., Morales-Muñiz, M., Pucher, E., Tournepiche, J.-F., Uerpmann, H.-P., Bălăşescu, A., Germonpré, M., Y. Günden, C., Hemami, M.-R., Moullé, P.-E., Öztan, A., Uerpmann, M., Walzer, C., Grange, T.*, Geigl, E.-M.* (2017) Taming the Late Quaternary phylogeography of the Eurasiatic wild ass through ancient and modern DNA. Plos one 12(4): e0174216. /doi.org/10.1371/journal.pone.0174216
Ottoni, C., Van Neer, W., De Cupere, B., Daligault, J., Guimaraes, S., Peters, J., Spassov, N., Prendergast, M.E., Boivin, N., Morales-Muniz, A., Bălăşescu, A., Becker, C., Benecke, N., Boronenanț, A., Buitenhuis, H., Chahoud, J., Crowther, A., Llorente, L., Manaseryan, N., Monchot, H., Onar, V., Osypińska, M., Putelat, O., Studer, J., Wierer, U., Decorte, R., Grange, T.*, Geigl, E.-M.* (2017) The paleogenetics of cat dispersal in the ancient world. Nature Ecology & Evolution 1, 0139. DOI: https://doi.org/10.1038/s41559-017-0139.
Grange, T., Brugal, J.-P., Flori, L., Gautier, M., Uzunidis, A., and Geigl, E.-M. (2018) The Evolution and Population Diversity of Bison in Pleistocene and Holocene Eurasia: Sex Matters. Diversity 10, 65; doi:10.3390/d10030065
Geigl, E.-M., Grange, T. (2018) Ancient DNA: The quest for the best. Mol Ecol Resour. 18:1185–1187. DOI: 10.1111/1755-0998.12931
Bennett, E.A., Crevecoeur, I., Viola, B., Derevianko, A.P., Shunkov, M.V., Grange, T., Maureille, B., Geigl, E.-M. (2019) Morphology of the Denisovan phalanx closer to modern humans than to Neandertals. Science Advances, 5:eaaw3950, DOI 10.1126/sciadv.aaw3950, DOI: 10.1126/sciadv.aaw3950
Stoetzel, E., Lalis, A., Nicolas, V., Aulagnier,S., Benazzou, T., Dauphin, Y., Abdeljalil El Hajraoui, M., El Hassani, A., Fahd, S., Fekhaoui, M., Geigl, E.-M., Lapointe, F.-J., Leblois, R., Ohler, A., Nespoulet, R., Denys, C.. Quaternary terrestrial microvertebrates from mediterranean northwestern Africa: State-of-the-art focused on recent multidisciplinary studies (2019) Quaternary Science Reviews 224: 105966. DOI: 10.1016/j.quascirev.2019.105966
Brunel, S., Bennett, E.A., Cardin, L., Garraud, D., Barrand Emam, H., Beylier, A., Boulestin, B., Chenal, F., Cieselski, E., Convertini, F., Dedet, B., Desenne, S., Dubouloz, J., Duday, H., Fabre, V., Gailledrat, E., Gandelin, M., Gleize, Y., Goepfert, S., Guilaine, J., Hachem, L., Ilett, M., Lambach, F., Mazière, F., Perrin, B., Plouin, S., Pinard, E., Praud, I., Richard, I., Riquier, V., Roure, R., Sendra, B., Thevenet, C., Thiol, S., Vauquelin, E., Vergnaud, L., Grange, T.*, Geigl, E.-M.*, Pruvost, M.* (2020) Ancient genomes from present-day France unveil 7,000 years of its demographic history. Proceedings of the National Academy of Sciences USA, 117(23):12791-12798, DOI:10.1073/pnas.1918034117.
Guimaraes, S., Arbuckle, B., Peters, J., Adcock, S., Buitenhuis, H., Chavin, H., Manaseryan, N., Uerpmann, H.-P., Grange, T.*, Geigl, E.-M.* (2020) Ancient DNA shows domestic horses were introduced in the southern Caucasus and Anatolia during the Bronze Age. Science Advances, 6: eabb0030, DOI: 10.1126/sciadv.abb0030
Geigl, E.-M. (2021) PCR et paléogénétique : pour le meilleur et pour le pire. PCR and paleogenetics: for best and for worst. Bulletin de l’Académie Nationale de Médecine, 205 (4) :389-395.
Balasse, M., Bellot-Gurlet, L., Dillmann, Geigl, E.-M., Jacob, J., Lebon, M., Le Hô, A.-S., Lefèvre, J.-C., Leroyer, C., Mathé, V., Merle, V., Mirabaud, S., Reiche, I. (2021) Service, expertise et archéométrie : état d’une réflexion collégiale. ArchéoSciences 2, n° 45-2, 81 – 88.
Bennett, E.A., Weber, J., Bendhafer, W., Champlot, S., Peters, J., Schwartz, G., Grange, T., Geigl, E.-M. (2022) The genetic identity of the earliest human-made hybrid animals, the kungas of Syro-Mesopotamia. Science Advances 8, eabm0218. DOI : 10.1126/sciadv.abm0218
Gorgé O., Bennett EA., Daligault J., Massilani D., Geigl E.-M.*, Grange T.* (2022) Analysis of ancient microbial DNA. Microbial Environmental Genomic, Methods Mol Biol 2605:103-131, doi: 10.1007/978-1-0716-2871-3_6
Giorgi, C., Bayle, G. Gourichon, L., Ben Dhafer, W., Geigl, E.-M. (2023) Un assemblage de restes de bovins au sein de l’établissement élitaire laténien de Palaiseau « Les trois mares » (Essonne). Revue archéologique d’Île-de-France, 13 : 179-212.Bennett, E.A., Parasayan, O., Prat, S., Péan, S., Crépin, L., Yanevich, A., Grange, T.,*, Eva-Maria Geigl, E.-M.* Genome sequences of 36,000 to 37,000 year-old modern humans at Buran-Kaya III in Crimea. (2023) Nature Ecology and Evolution 7, 2160–2172. https://doi.org/10.1038/s41559-023-02211-9
Jamieson A, Carmagnini A, Howard-McCombe J, Doherty S, Hirons A, Dimopoulos E, Lin AT, Allen R, Anderson-Whymark H, Barnett R, Batey C, Beglane F, Bowden W, Bratten J, De Cupere B, Drew E, Foley NM, Fowler T, Fox A, Geigl EM, Gotfredsen AB, Grange T, Griffiths D, Groß D, Haruda A, Hjermind J, Knapp Z, Lebrasseur O, Librado P, Lyons LA, Mainland I, McDonnell C, Muñoz-Fuentes V, Nowak C, O’Connor T, Peters J, Russo IM, Ryan H, Sheridan A, Sinding MS, Skoglund P, Swali P, Symmons R, Thomas G, Trolle Jensen TZ, Kitchener AC, Senn H, Lawson D, Driscoll C, Murphy WJ, Beaumont M, Ottoni C, Sykes N, Larson G, Frantz L. (2023) Limited historical admixture between European wildcats and domestic cats. Curr Biol. 33(21):4751-4760.e14. doi: 10.1016/j.cub.2023.08.031
Parasayan, O., Laurelut, C., Bole, C., Bonnabel, L., Corona, A., Domenech-Jaulneau, C., Paresys, C., Richard, I., Grange, T.*, Geigl, E.-M.* (2024) Late Neolithic collective burial reveals admixture dynamics during the third millennium BCE and the shaping of the European genome Sci. Adv. 10, eadl2468 (2024). Doi: 10.1126/sciadv.adl2468
Özkan M, Gürün K, Yüncü E, Vural KB, Atağ G, Akbaba A, Fidan FR, Sağlıcan E, Altınışık EN, Koptekin D, Pawłowska K, Hodder I, Adcock SE, Arbuckle BS, Steadman SR, McMahon G, Erdal YS, Bilgin CC, Togan İ, Geigl EM, Götherström A, Grange T, Özer F, Somel M. (2024) The first complete genome of the extinct European wild ass (Equus hemionus hydruntinus). Mol Ecol. 2024 Jul 1:e17440. doi: 10.1111/mec.17440.
Kaptan D, Atağ G, Vural KB, Morell Miranda P, Akbaba A, Yüncü E, Buluktaev A, Abazari MF, Yorulmaz S, Kazancı DD, Küçükakdağ Doğu A, Çakan YG, Özbal R, Gerritsen F, De Cupere B, Duru R, Umurtak G, Arbuckle BS, Baird D, Çevik Ö, Bıçakçı E, Gündem CY, Pişkin E, Hachem L, Canpolat K, Fakhari Z, Ochir-Goryaeva M, Kukanova V, Valipour HR, Hoseinzadeh J, Küçük Baloğlu F, Götherström A, Hadjisterkotis E, Grange T, Geigl EM, Togan İZ, Günther T, Somel M, Özer F. The Population History of Domestic Sheep Revealed by Paleogenomes. Mol Biol Evol. 2024 Oct 4;41(10):msae158. doi: 10.1093/molbev/msae158. PMID: 39437846; PMCID: PMC11495565.
Book chapters
Eckardt-Schupp, F., Geigl, E.-M., Ahne, F., Siede, W.: Is mismatch repair involved in UV-inducible mutagenesis in yeast? in: DNA Replication and Mutagenesis. (Eds.: R.E. Moses, W.C. Summers). Washington: ASM, 355-361, 1988
Leib-Mösch, C., Bachmann, M., Geigl, E.-M., Brack-Werner, R., Werner, T., Erfle, V., Hehlmann, R.: Expression of S71-related sequences in human cells. Haematol. and Blood Transfusion Vol. 35, p. 256-259 in Modern Trends in Human Leukemia IX, R. Neth et al. (Eds.), Springer Verlag Berlin Heidelberg 1992
Geigl, E.-M. and Pruvost, M. : Plea for a multidisciplinary approach to the study of Neolithic migrations : the analysis of biological witnesses and the input of palaeogenetics. In « Colonisation,, Migration, and Marginal Areas. A Zooarchaeological Approach. » M. Mondini, S. Munoz and S. Wickler (eds), Oxbow Books, p. 10-19, 2004.
Geigl, E.-M. : Why ancient DNA research needs taphonomy. In « Biosphere to Lithosphere: new studies in vertebrate taphonomy » T.O’Connor (ed.), Oxbow Books., p. 79-86, 2005.
Geigl, E.-M.: The domestication of cattle: Insights from a joint archaeozoological-palaeogenetical venture. In “Between Sand and Sea. The Archaeology and Human Ecology of Southwestern Asia”. N.J. Conard, P. Drechsler and A. Morales (eds.), Kerns Verlag Tübingen, p. 69-90, 2011.
Geigl, E.-M. Lima de Guimaraes, S., Liesau, C. Palaeogenetic analysis of bovine remains from Camino de las Yeseras and Humanejos. In “Yacimientos Calcoliticos con campaniforme de la region de Madrid: nuevos estudios ». Blasco, C., Liesau, C., and Rios, P. (eds.). Universidad Autonoma de Madrid, Madrid, p. 199-210, 2011.
Geigl, E.-M., Champlot, S., de Lima Guimaraes, S., Bennett, E.A., Grange, T. (2013) Molecular Taphonomy of Spy: DNA Preservation in Bone Remains. In Rougier, H. & Semal, P., (eds.) Spy cave. 125 years of multidisciplinary research at the Betche aux Rotches (Jemeppe-sur-Sambre, Province of Namur, Belgium), Volume 1. Anthropologica et Præhistorica, 123/2012. Brussels, Royal Belgian Institute of Natural Sciences, Royal Belgian Society of Anthropology and Praehistory & NESPOS Society, pp. 371-380.
Bennett, E.A., Cardin, L., Geigl, E.-M. (2013) Etude paléogénétique des momies d’Antinoé conservées au Musée des Beaux Arts de Lille. In « Les momies d’Antinoé ». Lintz, Y., Coudert, M. (eds.), p. 99-102
Geigl, E.-M., and Grange, T. Taphonomie de l’ADN ancien. (2014) In « Manuel de Taphonomie ». C. Denys et M. Patou-Mathis (eds), Editions errance, Collection archéologiques, p. 147-164.
Geigl, E.M. (2015) La paléogénétique et paléogénomique. In “Messages d’os”. M. Balasse, J.-P. Brugal, Y. Dauphin, E.-M. Geigl, C. Oberlin, I. Reiche (eds), Editions des Archives contemporaines, p. 429-431
Geigl, E.M. and Grange, T. (2015) Les stratégies et enjeux de l’analyse de l’ADN ancien. In “Messages d’os”. M. Balasse, J.-P. Brugal, Y. Dauphin, E.-M. Geigl, C. Oberlin, I. Reiche (eds), Editions des Archives contemporaines, p. 433-452
Pruvost,M. and Geigl, E.-M. (2015) « Apport de la paléogénétique à l’étude des processus de domestication animale . In “Messages d’os”. M. Balasse, J.-P. Brugal, Y. Dauphin, E.-M. Geigl, C. Oberlin, I. Reiche (eds), Editions des Archives contemporaines, p. 469-485
Geigl, E.M. and Grange, T. (2015) Le génome des lignées humaines archaïques. In “Messages d’os”. M. Balasse, J.-P. Brugal, Y. Dauphin, E.-M. Geigl, C. Oberlin, I. Reiche (eds), Editions des Archives contemporaines, p. 501-520
Eva-Maria Geigl, Shirli Bar-David, Albano Beja-Pereira, E. Gus Cothran, Elena Giulotto, Halszka Hrabar, Tsendsuren Oyunsuren, Mélanie Pruvost (2016): Genetics of wild equids. In “Wild Equids: Ecology, Conservation, and Management”, J. Ransom and P. Kazcensky (eds), p. 87-104
Geigl, E.-M., Guimaraes, S., Stoetzel, E., Fernandez-Jalvo, Y., Nespoulet, R., Denys, C., Grange, T. (2016) Analyse de la préservation de l’ADN dans les ossements des rongeurs de Témara, Maroc, en remontant dans le temps. In «Approche intégrative de la 6ème extinction : influence de l’installation des hommes modernes au Maroc sur l’évolution de la biodiversité des petits vertébrés terrestres ». Travaux de l’institut Scientifique de l’université Mohammed V de Rabat, Série générale, N° 8, 43-51
Geigl, E.-M., Bennett, E.A., Grange, T. (2016) Analyse de la degradation de l’ADN dans une phalange de la Dame du Cavillon. In “La grotte du Cavillon”. H. de Lumley (ed), CNRS Editions, p. 969-975.
Bennett, E. A., Gorgé, O., Grange, T., Fernández-Jalvo, Y. and Geigl, E.-M. (2016) Coprolites, Paleogenomics and Bone Content Analysis. In « Azokh Cave and the Transcaucasian Corridor». Yolanda Fernández-Jalvo, Tania King, Levon Yepiskoposyan, and Peter Andrews (eds), Springer, Vertebrate Paleobiology and Paleoanthropology Series, E. Delson and E.J. Sargis (eds), p. 271-286 DOI 10.1007/978-3-319-24924-7 (peer reviewed article)
Geigl, E.-M. & Grange, T. (2018) Of Cats and Men: Ancient DNA Reveals How the Cat Conquered the Ancient World. Charlotte Lindqvist and Om P. Rajora (eds.), Paleogenomics, Population Genomics [Om P. Rajora (Editor-in-Chief)], Springer International Publishing AG, part of Springer Nature 2018. DOI https://doi.org/10.1007/13836_2018_26 (peer reviewed article)
Geigl, E.-M. & Grange, T. (2019) Using palaeogenetics to unravel the impact of humans on animal populations in the past. In Animals: Cultural Identifiers in Ancient Societies? J. Peters, G. McGlynn & V. Goebel (eds)., Verlag Marie Leidorf (VML) GmbH, Rahden/Westf. p.131-137
Brunel, S., Bennett, E.A., Grange, T., Geigl, E.M., Pruvost, M. (2019) Etude paléogénomique des ossements du dolmen de Saint-Eugène. In Le dolmen de Saint-Eugène : autopsie d’une sépulture collective néolithique. Jean Guilaine., 1 vol., Archives d’écologie préhistorique, Toulouse, France. pp.339-345, 2020, 9782358420266 (pp. 405)
Liesau, C., Ríos, P., Vega, J., Blasco, C., Menduiña, R., de los Ángeles de Chorro, M., Cabrera, C., Geigl, E.-M., and Arteaga, C. (2022) The Bovine Deposits from the Chalcolithic Ditched Enclosure of Camino de las Yeseras (Madrid, Spain). In Wright, L. and Ginja, C. (eds) Cattle and People Interdisciplinary Approaches to an Ancient Relationship. Archaeobiology, 4, Series Editors Whitcher Kansa, S. and Lev-Tov, J., Lockwood Press, Columbus pp. 241-261.
Geigl, E.-M. (2023) Palaeogenetics and Palaeogenomics to Study the Domestication of Animals. In A. Mark Pollard, Ruth Ann Armitage, Cheryl A. Makarewicz (eds). Handbook of Archaeological Sciences DOI: 10.1002/9781119592112.ch33
PUBLICATIONS FOR THE GENERAL PUBLIC
1 Geigl, E.-M.: L’émergence de la paléogénétique. Biofutur 164:28-34, 1997
3 Geigl, E.-M., Grange, T., Maureille, B. : Le génome néandertalien. Encyclopaedia Universalis, « La Science au présent 2011 », p. 133-140
4 Geigl, E.-M. : Le métier du paléogénéticien. Archéothéma, Hors-série « Les métiers de l’archéologie » 2012, p. 58-59
5 « Mondes polaires – Hommes et biodiversités, des défis pour la science » (2012) R. Chenorkian, M. Raccurt (eds), Cherche-Midi
6 Geigl, E.-M. (2012) Les relations entre les humains et les animaux à la lumière de la génétique. L’archéologie au laboratoire. Coédition INRAP, Cité des Sciences et de l’Industrie Paris et éditions « La Découverte », p.61-74
7 Geigl, E.-M., Grange, T. (2013) Du potentiel mais des défis méthodologiques à relever. Biofutur 349, 24-27.
8 Geigl, E.-M. (2013) Conserver ou ressusciter les espèces en voie de disparition ? Biofutur 34, 32-34.
9 Pruvost, M., Massilani, D., Geigl, E.-M. (2013) La domestication retracée par l’ADN ancien. Biofutur 349, 36-39.
10 Geigl, E.-M. (2013) Quand étudier le génome des lignées humaines éteintes devient possible. Biofutur 349, 45-49.
11 Geigl, E.-M. (2015) L’apport de la paléogénétique et paléogénomique à l’archéologie. Nouvelles d’Archéologie 138, 10-14.
12 Geigl, E.-M., Bennett, E.A., Grange, T. (2015) Tracing the origin of our species through paleogenomics. BIO Web of Conferences 4, 00005 (2015) DOI: 10.1051/bioconf/20150400005. Owned by the authors, published by EDP Sciences, 2015
13 Geigl, E.-M. and Grange, T. (2016) La paléogénomique, pour une lecture du passé au présent. « L’empreinte du vivant » D. Joly, D. Faure, S. Salamitou (eds)., Editions Cherche Midi, p.151-165.
14 Geigl, E.-M. et Grange, T. (2017) Biodiversité passée et paléogénomique. « Les Big Data à Découvert », Bouzeghoub, M. et Mosseri, R. (eds), CNRS Editions, p. 228 – 229
15 Geigl, E.-M. et Grange, T. (2017) How cats conquered the Ancient world: a 9,000-years DNA tale. The Science Breaker. doi.org/10.25250/thescbr.brk062
16 Geigl, E.-M. (2017) Le peuplement de l’Europe vu par la paléogénomique. In « L’archéologie des migrations », Garcia D. and Le Bras, H. (eds.), La Découverte, Inrap, p. 67 – 80.
17 Geigl, E.-M. et Grange, T. (2017) « Comment le chat a conquis le monde ». Dans « De la Préhistoire à nos jours : Le Chat – Comment il a conquis le monde » Historia, Décembre 2017.
18 Grange, T. et Geigl, E.-M. (2018) « Grâce à la domestication, le chat a gagné le monde ». La Recherche, 531 :65-68
19 Geigl, E.-M. (2018) L’évolution des mammouths vue par la paléogénomique. In : Mémoire de Mammouth (244 p): [Exposition Musée national de Préhistoire – Les Eyzies de Tayac 29 juin – 12 novembre 2018] / Commissariat de l’exposition : Catherine Cretin, Stéphane Madelaine. Comité scientifique : Gennady Boeskorov, Peggy Bonnet-Jacquement, Jean-Jacques Cleyet-Merle, Philippe Fosse, Frédéric Plassard. Les Eyzies-de-Tayac, Musée national de Préhistoire. p. 31-34
20 Geigl, E.-M. (2018) La paléogénétique en tant qu’approche archéométrique au cours des 30 dernières années – Paleogenetics as Archeometrical approach over the last 30 years. ArcheoSciences, revue d’archéométrie, 42(1) : 95-104.
21 Grange, T. et Geigl, E.-M. (2019) « Grâce à la domestication, le chat a gagné le monde ». La Recherche – Les Essentiels, n° 30, p. 82-85
22 Geigl, E.-M. & Grange, T. (2019) Changements climatiques : 150 000 ans d’évolution des populations de bisons en Europe. In « 101 sécrets de l’ADN », Faure, D., Joly, D. Salamitou, S. (eds). Editions CNRS, p. 264-266.
23 Geigl, E.-M. (13/01/2020) « Une petite phalange réécrit l’histoire évolutive des humains », The Conversation https://theconversation.com/une-petite-phalange-reecrit-lhistoire-evolutive-des-humains-123156
24 Geigl, E.-M. (2020) « Le Peuplement de la France à la Protohistoire grâce à la paléogénomique ». Archéologia n° 589, 16-17.
25 Geigl, E.-M. (2020) “Qui a habité en France ces 9 000 dernières années ? » The Conversation https://theconversation.com/qui-a-habite-en-france-ces-9-000-dernieres-annees-139769
26 Geigl, E.-M. (2021) “Contribution de la paléogénétique à l’archéologie » In C. Carpentier, R.-M. Arbogast & Ph. Kuchler (dir.), Bioarchéologie : minimums méthodologiques, référentiels communs et nouvelles approches : actes du 4e séminaire scientifique et technique de l’Inrap, 28-29 nov. 2019, Sélestat. https://doi.org/10.34692/gdqj-7g88
27 Geigl, E.-M. (2023) “Crimea’s place in Europe 36,000 years ago: a tale of ancient genomes” https://ecoevocommunity.nature.com/manage/posts/211298
27 Geigl, E.-M., Grange, T. (2023) “Homo sapiens : comment deux crânes réécrivent l’histoire de son apparition en Europe – Nouvelle recherche » https://theconversation.com/homo-sapiens-comment-deux-cranes-reecrivent-lhistoire-de-son-apparition-en-europe-nouvelle-recherche-216195 ; https://theconversation.com/skulls-in-ukraine-reveal-early-modern-humans-came-from-the-east-216553
28 Geigl, E.-M., Parasayan, O., Grange, T. (2024) « Une tombe collective de 4 500 ans révèle son secret : la dernière étape de la formation du génome européen » https://theconversation.com/une-tombe-collective-de-4-500-ans-revele-son-secret-la-derniere-etape-de-la-formation-du-genome-europeen-232627
29 Geigl, E.-M., Parasayan, O., Grange, T. (2024) « A 4,500-year-old collective tomb in France reveals its secret – the final stage in the formation of the European genome » https://theconversation.com/a-4-500-year-old-collective-tomb-in-france-reveals-its-secret-the-final-stage-in-the-formation-of-the-european-genome-233382
30 Mattei, J., Grange, T., Geigl, E.-M. (2024) Le changement du rapport entre chat et humain au cours des 10000 dernières années. Revue semestrielle de droit animalier (RSDA) https://www.revue-rsda.fr/articles-rsda/7579-le-changement-du-rapport-entre-chat-et-humain-au-cours-des-10000-dernieres-annees
- Jeanne Mattei « Paléogénomique de la domestication des chats » soutenue le 20 décembre 2023
- Oguzhan Parasayan « Genomic evolution of of ancient French populations » soutenue le 20 décembre 2022
- Wejden Bendhafer : « Paléogénomique de l’évolution des Bovina et l’impact sur la domestication des bovins». Soutenance de thèse le 20/12/2021.
- Caitlin Martin : « The evolution of and selection upon the human genome since the invention of agriculture: a historical perspective on diseases”; en cours depuis Octobre 2020
- Samantha Brunel : « Analyse paléogénétique du peuplement de la France ». Soutenance de thèse le 14/11/2018.
- Diyendo Massilani : « Paléogénomique des Bovina et de leur domestication ». Soutenance de thèse le 6/7/ 2016.
- Olivier Gorgé : «Diagénèse de l’ADN bactérien et analyses métagénomiques de pathologies bactériennes du passé ». Soutenance de thèse le 13/12/2016.
- Nathalie Côté : « Apports de la paléogénétique à l’étude des helminthes garstro-interstinaux anciens». Soutenance de thèse le 16/12/2015 .
- Sophie Champlot : « Colonisation et domestication de l’Europe et du pourtour méditerranéen », Ecole Doctorale « Gènes, Génomes et Cellules », soutenue le 13/7/2010.
Collaborations scientifiques avec plus de 60 chercheurs nationaux et internationaux de 20 pays, surtout des archéologues, archéozoologues, paléoanthropologues.
Financements de l’équipe depuis 2010 :
2021-2024 Projet européen WIDESPREAD-05-2020 – Twinning: « Mapping The Neolithic Expansion In The Mediterranean: A Scientific Collective To Promote Archaeogenomics And Evolutionary Biology Research In Turkey” (H2020-WIDESPREAD-2020-5; CSA 952317; NEOMATRIX
2018-2021 “Paléogénomique de la domestication des bovins pour enrichir les stratégies durables de sélection (PATH2BOS) », ANR, coordinateur : Thierry Grange
2016-2021 “ Etude Génétique de la population Française (FROGH)“, ANR, coordinateur : Christian Dina, Institut du Thorax, UMR 1087/ UMR 6291, Nantes. Equipe « Epigénome & Paléogénome » : « Paléogénomique de la population médiévale de l’Ouest de la France ».
2015-2019 “Genetic characterization of Ancestral French populations using ancient DNA (ANCESTRA)” ANR JCJC – ANR-15-CE27-0001; Coordinatrice: Mélanie Pruvost, Institut Jacques Monod, Paris.
2014-2016 “The origins of Equid domestication”, National Science Foundation (NSF) Prime Award N° BCS-1311551; Coordinateur: Benjamin Arbuckle, University of Chapel Hill, North Carolina, USA. « Epigénome & Paléogénome » : “Paleogenetics of the domestication of horses in Anatolia and the Caucasus”
2010-2014 “Influence de l’installation des hommes modernes au Maroc sur l’évolution de la biodiversité des petits vertébrés terrestres (MOHMIE)“ ; Agence Nationale de Recherche (ANR) n° 2009 PEXT 004 05; Coordinatrice : C. Denys, Muséum National d’Histoire Naturelle. « Epigénome & Paléogénome » : “Paléogénétique des rongeurs au cours des derniers 120,000 ans”[/vc_column_text][/vc_tta_section][vc_tta_section title=”Actualités” tab_id=”1645025044634-e8b93c72-ce32″][vc_column_text]
Vidéo on the Université Paris Cité Youtube channel: Paléogénomique : la double hélice comme machine à explorer le temps
November 19th to December 16th 2024: [Symphosium & Exhibition]The peopling of France and the origins of the agricultural lifestyle
- Free registration but mandatory to eva-maria.geigl@ijm.fr
- AGRIPOP exhibition
23 octobre 2023 – Nouvel article publié dans Nature Ecology and Evolution : Genome sequences of 36-37,000 year-old modern humans at Buran-Kaya III in Crimea
10 mars 2023 – Conférence au Collège de France ” L’évolution des populations humaines et animales : une histoire de migrations et métissages” présenté dans le cadre du cours “L’histoire de l’humanité vue sous l’angle de la paléogénomique” donné par Lluis Quinatana-Murci.
A new study by the Grange/Geigl team published in Science Advances on 14 January 2022!
- Before the introduction of the domestic horse in Mesopotamia, valuable equids were being harnessed to ceremonial or military four wheeled wagons and used as royal gifts, but their true nature remained unknown.
- According to a palaeogenetic study, these prestigious animals were the result of a cross between a domestic donkey and a wild ass from Syria, now extinct.
- This makes them the oldest example of an animal hybrid produced by humans.
- Communiqué de presse
Fête de la science 2021 – Conférence “La génétique du peuplement de l’Europe depuis l’âge des glaces”
Communications de notre recherche par le CNRS :
https://archives.cnrs.fr/insb/article/2016/e-m-geigl
https://lejournal.cnrs.fr/articles/comment-le-chat-a-conquis-le-monde
https://www.cnrs.fr/fr/7-000-ans-dhistoire-demographique-en-france
https://www.cnrs.fr/fr/avant-les-chevaux-des-hybrides-danes-produits-pour-faire-la-guerre