Regulation of Cell-Fate Specification
Jérôme COLLIGNON
We are studying the role of a secreted protein, called NODAL, in the specification of cell lineages during embryogenesis. This has led us to study the role of this molecule in tumour progression too.
Keywords: Mouse embryo, embryonic stem cells, human melanoma, ACTIVIN/NODAL signalling, regulation of gene expression, enhancers, pluripotency
+33 (0)1 57 27 81 08 Contact
We are studying the role of a secreted protein, called NODAL, in the specification of cell lineages during embryogenesis. This has led us to study the role of this molecule in tumour progression too.
Studies in the mouse embryo have shown that the specification of cell fates involves several signalling pathways. These pathways are also involved in the maintenance and renewal of tissues in the adult, functions that are implicated in many diseases. The study of their involvement and interactions during embryonic development, when the organism is still relatively simple, is therefore particularly relevant for understanding their role later, in more complex contexts.
NODAL is a member of the TGFß family. Its signal is transmitted through the so-called ACTIVIN/NODAL signalling pathway, which depends on the cytoplasmic factors SMAD2 and 3. The Nodal gene is required for the establishment of embryonic axes. It is also required for the specification and maintenance of various cell identities, both embryonic and extra-embryonic. Many of these cell types have in common that they are motile. The Nodal locus has been the subject of numerous studies, its regulatory sequences have been identified and characterised, and it has become a model for understanding how various influences (TGFß, WNT and NOTCH signals, pluripotency factors, etc.) are integrated and cause a gene to be expressed in a certain cell at a certain time. In adults, Nodal is expressed in tissues that are periodically renewed or remodelled under the control of hormonal stimuli, such as the endometrium and the mammary gland. In humans, NODAL expression has been detected in tumour cells of different types of cancers and has been associated with their plasticity and invasive behaviour, suggesting that its role in this context reflects what is known about its functions in adult and embryonic tissues.
We are currently using a combination of genetic, molecular biology, transcriptomic and imaging approaches to study the role of NODAL during specification and differentiation of early cell lineages, both in the mouse embryo and in vitro culture models based on the use of embryonic stem cells. The study of these in vitro models is carried out in collaboration with biophysicists who contribute their expertise in microfluidics, quantitative data processing and modelling. Finally, we use cell biology, genetics, biochemistry, transcriptomics and mass spectrometry approaches to study the mechanisms by which an antisense transcript of the NODAL gene promotes tumour progression in a human melanoma cell model.
Study of the role of Nodal at preimplantation stages
Nodal expression is first detected in the inner cell mass (ICM) of the early blastocyst at embryonic day 3.5 (E3.5) (Granier et al., 2011; Takaoka et al., 2006; Takaoka et al., 2017). At E3.75, when the ICM is giving rise to the pluripotent epiblast (Epi) and to the primitive endoderm (PrE), an extra-embryonic layer, Nodal expression persists in the Epi but becomes stronger in PrE cells. At E4.5, when the embryo implants, Nodal expression is downregulated in the PrE but gains in strength throughout the Epi. The activity of the ACTIVIN/NODAL signalling pathway at these stages was found to track Nodal expression quite closely (Granier et al., 2011).
The earliest defect described in Nodal-/- embryos so far is their inability to activate the expression of Lefty1, a feedback antagonist of NODAL, in ICM and PrE cells between E3.5 and E4.5 (Takaoka et al., 2017). Nodal-/- E4.5 embryos nevertheless comprise the three cell layers that normally constitute an implanting blastocyst. It is only after implantation that their signature developmental defects, the failure to establish specific embryonic and extra-embryonic cell identities, to establish an AP axis and to gastrulate, have been extensively characterized (Robertson 2014). Apart from Lefty1, other possible targets of NODAL at preimplantation stages have been identified in studies of embryonic stem cells (ESCs), which are representative of early Epi cells (Galvin et al., 2010; Mullen et al., 2011). Given the prominence of some of these targets, which include transcription factors and components of signalling pathways, the absence of a preimplantation phenotype may seem surprising. One possible reason for this apparent lack of early defects is the presence at these stages of TGFb-related ligands of zygotic (ACTIVIN) and uterine (ACTIVIN, NODAL) origins susceptible to compensate for the absence of zygotic NODAL (Papanayotou & Collignon, 2014). Evidence that ACTIVIN/NODAL signalling is active in the embryo well before zygotic Nodal expression begins, is consistent with this possibility (James et al., 2005).
In order to reveal the respective contributions of NODAL and ACTIVIN to the formation of the blastocyst we have undertaken a comparison of the phenotypes of embryos exposed to various inductive or inhibitory treatments with that of freshly dissected or ex-utero cultured Nodal-/- embryos.
This project received funding from the Émergence program (IDEX Université Paris Cité), Fondation ARC, the doctoral school BioSPC, the EUR G.E.N.E and the Labex Who Am I?
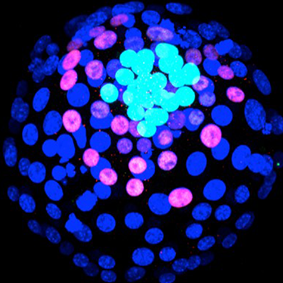
E4.0 mouse embryo. Dark blue nuclei are in trophectoderm cells, pink nuclei are in primitive endoderm cells (stained with a SOX17 antibody), light blue nuclei are in epiblast cells (SOX2 antibody).
Study of the mechanisms underlying the patterning of the primitive streak
We aim to understand how the primitive streak (PS), the formation of which marks the beginning of gastrulation, is patterned in the mouse. The PS appears at the edge of the epiblast cup at a position corresponding to the posterior side of the embryo, and extends gradually towards its distal tip. Cells emerging early on from the proximal/posterior region of the PS will migrate proximally to form extra-embryonic mesoderm or laterally to form embryonic mesoderm, whereas cells emerging from its distal/anterior region will form axial mesoderm and definitive endoderm. Complete and partial loss-of-function mutations showed that three secreted ligands, WNT3, BMP4 and NODAL, are critical for the formation and the patterning of the PS. These studies led to a model of PS patterning where opposing gradients of NODAL and BMP4 signalling govern the allocation of cell fates along its anterior/posterior (A/P) axis, with the highest BMP signalling promoting extra-embryonic mesoderm in the posterior PS, and the highest NODAL signalling promoting definitive endoderm in the anterior PS (Tam et al., 2006; Arnold & Robertson 2007). While there is evidence of such a gradient of BMP signalling (Yamamoto et al., 2009), it is unclear for NODAL signalling (Camacho-Aguilar & Warmflash, 2020). Evidence from zebrafish suggests that a long exposure to NODAL signalling does promote anterior PS fates (Hill, 2018), but in the mouse embryo it is difficult to ascertain the relationship between ligand exposure and cell-fate allocation. Furthermore, the mutual interdependency of the expression of Wnt3, Bmp4 and Nodal, and their earlier roles in embryo patterning, make it challenging to disentangle, for example, the effect of the lack of NODAL from those of a lack of BMP4 and WNT3 in Nodal-/- embryos patterning defects. Finally, the small amount of biological material in one embryo makes it difficult to apply biochemical methods to study how cell-fate decisions integrate these 3 signals. However, the recent development of methods that use pluripotent cells to recapitulate gastrulation in vitro, thus allowing greater control over the conditions under which patterning takes place, now makes such investigations possible.
To compare events taking place in vitro with those actually taking place in the mouse embryo, our collaborator Benoit Sorre (UMR168 Institut Curie/CNRS) adapted a method he originally developed with human embryonic stem cells (hESCs) (Warmflash et al., 2014) to the use of mouse pluripotent stem cells representative of the pre-gastrulation epiblast, the EpiLCs (Plouhinec et al., 2021). This 2D in vitro model of gastrulation, or 2D mouse gastruloïds (m2DGas), has many advantages. It is simple to set up, multiple colonies can be generated at once, and it is amenable to imaging and quantification. We found, like others (Morgani et al., 2018), that m2DGas, reproducibly self-organize when exposed to BMP4, forming within 48H a PS that then gives rise to a ring of mesoderm cells surrounding a core of ectoderm. We are now using an array of approaches (Immunofluorescence, in situ hybridization, RT-qPCR, single-cell RNAseq, gene regulatory network inference, proximity ligation assay) to study in this model the mechanisms underlying the patterning of the PS.
This project has been supported by INCa, and the doctoral school BioSPC. It is currently supported by funding from the ANR.
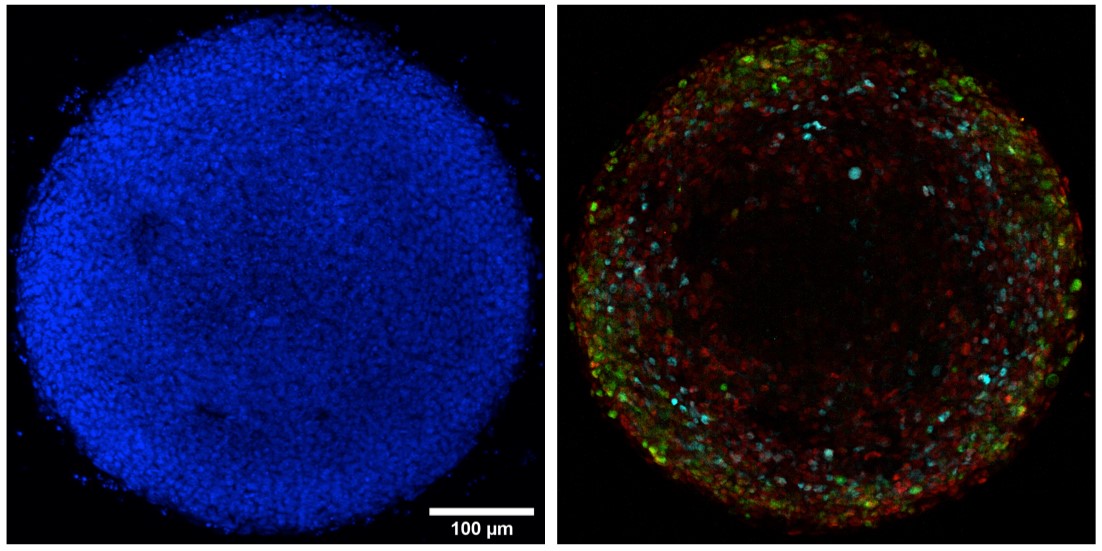
500 μm diameter EpiLC colony after 36h BMP4 stimulation. On the left, Hoechst (blue) labelling of nuclei allows visualization of all cells, on the right,,immunofluorescence labelling allows identification of primitive streak and nascent mesoderm cells (BRA in red), extra-embryonic mesoderm cells (CDX2 in green) and proximal primitive streak cells (FOXA2 in cyan).
Study of the role of the NODAL antisens LADON in human melanoma tumour progression
The involvement of NODAL in tumour progression and metastasis was first described in melanoma cell lines (Topczewska et al., 2006). While evidence supporting such a role for this ligand was obtained in other cancer types, its actual contribution to metastasis in melanoma was challenged (Donovan et al., 2017; Hooijkaas et al., 2011), notably because its expression was not always detected. A review of available data on the expression of NODAL in cancer cells suggested the existence of distinct splice variants (Strizzi et al., 2012), all including the second exon of the gene but sometimes, notably in melanoma cell lines, lacking its third (and last) exon and therefore the capacity to produce a functional ligand. This observation provided an explanation for reported discrepancies between different studies regarding NODAL expression levels in these cell lines, but raised also the possibility of a ligand-independent function of the gene.
We thus used genome editing to delete NODAL exon2 in the A375 melanoma cell line, a recognized melanoma cell culture model previously described as NODAL-expressing (Topczewska et al., 2006). The mutation drastically reduced the capacity of these cells to acquire an invasive phenotype. However, the exogenous inhibition or stimulation of ACTIVIN/NODAL signalling had no effect on the behaviour of A375 cells, and we could not detect actual NODAL expression in these cells, implying that neither the NODAL ligand nor its mRNA played a part in this phenotype. We found instead that the mutation disabled the expression of a 1750 nucleotide-long natural antisense transcript overlapping with the entire exon2 of NODAL (Fig.1). Originally identified in glioblastoma, it had thus far been largely unnoticed. We called this transcript LADON and confirmed that this is what previous studies mistakenly identified as NODAL expression in melanoma cell lines, and in some other cancer cell lines too.
There are multiple examples of the implication of long non-coding RNAs (lncRNAs) in tissue physiology and in the misregulation of cellular processes, which often result in cancer (Huarte 2015; Iyer et al., 2015; Schmitt & Chang). This is the reason for the interest lncRNAs currently elicit as biomarkers for diagnostic and prognosis, but also as potential targets for therapy or as tools to correct defective gene expression underlying cancer and other pathologies (Wahlestedt 2013; Slaby et al., 2017). While we have characterised some of the events that depend on the expression of LADON in melanoma cells we do not know how it is actually triggering them. Our objectives are therefore to identify LADON’s direct targets and to characterize how it is controlling their expression.
This project received funding from INCa and GEFLUC
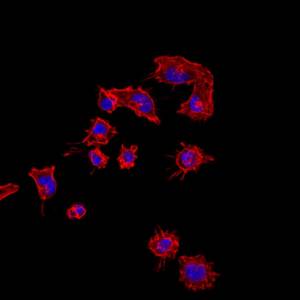
Melanoma cells of the A375 lineage, whose acquisition of invasive behaviour depends on the expression of the LADON transcript. The nuclei are in blue and the cytoskeleton in red.
Members
 Mariama Dianké DANSOKHO, Intern, COLLIGNON LAB
Mariama Dianké DANSOKHO, Intern, COLLIGNON LAB Annie DUTRIAUX, Biology engineer, COLLIGNON LAB+33 (0)1 57 27 81 10, room 502B
Annie DUTRIAUX, Biology engineer, COLLIGNON LAB+33 (0)1 57 27 81 10, room 502B Domenico FLAGIELLO, Assistant Professor, COLLIGNON LAB+33 (0)1 57 27 80 86, room 502B
Domenico FLAGIELLO, Assistant Professor, COLLIGNON LAB+33 (0)1 57 27 80 86, room 502B Futura GATTOBIGIO, PhD student, COLLIGNON LAB+33 (0)1 57 27 81 10, room 502B
Futura GATTOBIGIO, PhD student, COLLIGNON LAB+33 (0)1 57 27 81 10, room 502B Gaël SIMON, Postdoctoral researcher, COLLIGNON LAB+33 (0)1 57 27 81 09, room 502B
Gaël SIMON, Postdoctoral researcher, COLLIGNON LAB+33 (0)1 57 27 81 09, room 502B
To contact a member of the team by e-mail: name.surname@ijm.fr
BMP signaling modulations control primitive streak patterning. Simon G, Plouhinec JL, Gilardi-Hebenstreit P, Sorre B*, Collignon J*. bioRxiv 2024.10.01.616050; doi: https://doi.org/10.1101/2024.10.01.616050
LADON, a natural antisense transcript of NODAL, promotes metastasis promotes tumour progression and metastasis in melanoma. Dutriaux A, Diazzi S, Bresesti C, Hardouin S, Deshayes F, Collignon J*, Flagiello D*. Non-Coding RNA. 2023 Nov 15;9(6):71. doi: 10.3390/ncrna9060071. PMID: 37987367
Dissecting signalling hierarchies in the patterning of the mouse primitive streak using micro-patterned EpiLC colonies. Plouhinec JL#, Simon G#, Vieira M, Collignon J*, Sorre B*. Stem Cell reports. 2022 Jul 12;17(7):1757-1771. doi: 10.1016/j.stemcr.2022.05.009. PMID: 35714597
A conserved role for non-neural ectoderm cells in early neural development. Cajal M, Creuzet S, Papanayotou C, Sabéran-Djoneidi D, Chuva de Sousa Lopes SM, Zwijsen A, Collignon J and Camus A. Development. 2014 Nov;141(21):4127-38. doi: 10.1242/dev.107425. Epub 2014 Oct 1. PMID: 25273086
Activin/Nodal signalling before implantation: setting the stage for embryo patterning. Papanayotou C and Collignon J. Philos Trans R Soc Lond B Biol Sci. 2014 Dec 5;369(1657):20130539. doi: 10.1098/rstb.2013.0539. PMID: 25349448
A novel Nodal enhancer dependent on pluripotency factors and Smad2/3 signaling conditions a regulatory switch during epiblast maturation. Papanayotou C, Benhaddou A, Camus A, Perea-Gomez A, Jouneau A, Mezger V, Langa F, Ott S, Sabéran-Djoneidi D, Collignon J. PLoS Biol. 2014 Jun 24;12(6):e1001890. doi: 10.1371/journal.pbio.1001890. eCollection 2014 Jun. PMID: 24960041
Clonal and molecular analysis of the prospective anterior neural boundary in the mouse embryo. Cajal M, Lawson KA, Hill B, Moreau A, Rao J, Ross A, Collignon J, Camus A. Development. 2012 Jan;139(2):423-36. doi: 10.1242/dev.075499. PMID: 22186731
Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo. Granier C#, Gurchenkov V#, Perea-Gomez A, Camus A, Ott S, Papanayotou C, Iranzo J, Moreau A, Reid J, Koentges G, Sabéran-Djoneidi D, Collignon J. Dev Biol. 2011 Jan 15;349(2):350-62. doi: 10.1016/j.ydbio.2010.10.036. Epub 2010 Nov 1. PMID: 21047506
Absence of Nodal signalling promotes precocious neural differentiation in the mouse embryo. Camus A, Perea-Gomez A, Moreau A and Collignon J. Dev Biol. 2006 Jul 15;295(2):743-55. doi: 10.1016/j.ydbio.2006.03.047. Epub 2006 Apr 7. PMID: 16678814
Initiation of gastrulation in the mouse embryo is preceded by an apparent shift in the orientation of the anterior-posterior axis. Perea-Gomez A, Camus A, Moreau A, Grieve K, Moneron G, Dubois A, Cibert C and Collignon J. Curr Biol. 2004 Feb 3;14(3):197-207. doi: 10.1016/j.cub.2004.01.030. PMID: 14761651
Neural crest cell recruitment and reprogramming as central drivers of embryonic limb regeneration. Laplace-Builhé B, Tejedor G, De La Cruz J, Barthelaix A, Marmigère F, Sapède D, Bahraoui S, Diouloufet L, Ventéo S, Collignon J, Jorgensen C, Djouad F. Proc Natl Acad Sci U S A. 2025 Dec 30;122(52):e2519994122. doi: 10.1073/pnas.2519994122.
BMP signaling modulations control primitive streak patterning. Simon G, Plouhinec JL, Gilardi-Hebenstreit P, Sorre B*, Collignon J*. bioRxiv 2024.10.01.616050; doi: https://doi.org/10.1101/2024.10.01.616050
LADON, a natural antisense transcript of NODAL, promotes metastasis promotes tumour progression and metastasis in melanoma. Dutriaux A, Diazzi S, Bresesti C, Hardouin S, Deshayes F, Collignon J*, Flagiello D*. Non-Coding RNA. 2023 Nov 15;9(6):71. doi: 10.3390/ncrna9060071. PMID: 37987367
Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi. Tomaino G, Pantaleoni C, Ami D, Pellecchia F, Dutriaux A, Barbieri L, Garbujo S, Natalello A, Tortora P, Frascotti G. Int J Mol Sci. 2023 Feb 20;24(4):4214. doi: 10.3390/ijms24044214. PMID: 36835627
Dissecting signalling hierarchies in the patterning of the mouse primitive streak using micro-patterned EpiLC colonies. Plouhinec JL#, Simon G#, Vieira M, Collignon J*, Sorre B*. Stem Cell reports. 2022 Jul 12;17(7):1757-1771. doi: 10.1016/j.stemcr.2022.05.009. PMID: 35714597
Whole embryo culture, transcriptomics and RNA interference identify TBX1 and FGF11 as novel regulators of limb development in the mouse. Tejedor G, Laplace-Builhé B, Luz-Crawford P, Assou S, Barthelaix A, Mathieu M, Kissa K, Jorgensen C, Collignon J*, Chuchana P*, Djouad F*. Sci Rep. 2020 Feb 27;10(1):3597. doi: 10.1038/s41598-020-60217-w. PMID: 32107392
Loss of Cubilin, the intrinsic factor-vitamin B12 receptor, impairs visceral endoderm endocytosis and endodermal patterning in the mouse. Perea-Gomez A, Cases O, Lelièvre V, Pulina MV, Collignon J, Hadjantonakis AK, Kozyraki R. Sci Rep. 2019 Jul 15;9(1):10168. doi: 10.1038/s41598-019-46559-0. PMID: 31308417
Evolutionary Gain of Dbx1 Expression Drives Subplate Identity in the Cerebral Cortex. Arai Y, Cwetsch AW, Coppola E, Cipriani S, Nishihara H, Kanki H, Saillour Y, Freret-Hodara B, Dutriaux A, Okada N, Okano H, Dehay C, Nardelli J, Gressens P, Shimogori T, D’Onofrio G, Pierani A. Cell Rep. 2019 Oct 15;29(3):645-658.e5. doi: 10.1016/j.celrep.2019.09.007. PMID: 31618633
A requirement for ZIC2 in the regulation of Nodal expression underlies the establishment of left-sided identity. Dykes IM, Szumska D, Kuncheria L, Puliyadi R, Chen CM, Papanayotou C, Lockstone H, Dubourg C, David V, Schneider JE, Keane TM, Adams DJ, Brown SDM, Mercier S, Odent S, Collignon J, Bhattacharya S. (2018). Sci Rep. 2018 Jul 11;8(1):10439. doi: 10.1038/s41598-018-28714-1. PMID: 29992973
Anteroposterior polarity and elongation in the absence of extraembryonic tissues and spatially localised signalling in Gastruloids, mammalian embryonic organoids. Turner DA, Girgin M, Alonso-Crisostomo L, Trivedi V, Baillie-Johnson P, Glodowski CR, Hayward PC, Collignon J, Gustavsen C, Serup P, Steventon B, Lutolf M, and Martinez Arias A. Development. 2017 Nov 1;144(21):3894-3906. doi: 10.1242/dev.150391. Epub 2017 Sep 26. PMID: 28951435
Paternal Insulin-like Growth Factor 2 (Igf2) Regulates Stem Cell Activity During Adulthood. Barroca V, Lewandowski D, Jaracz-Ros A, Hardouin SN. EBioMedicine. 2017 Feb;15:150-162. doi: 10.1016/j.ebiom.2016.11.035. Epub 2016 Dec 3. PMID: 28007480
An evolutionary, structural and functional overview of the mammalian TEAD1 and TEAD2 transcription factors. Landin-Malt A, Benhaddou A, Zider A, Flagiello D. Gene. 2016 Oct 10;591(1):292-303. doi: 10.1016/j.gene.2016.07.028. Epub 2016 Jul 14. PMID: 27421669
Notch activation interferes with cell fate specification in the gastrulating mouse embryo. Souilhol C#, Perea-Gomez A#, Camus A, Beck-Cormier S, Vandormael-Pournin S, Escande M, Collignon J and Cohen-Tannoudji M. Development. 2015 Nov 1;142(21):3649-60. doi: 10.1242/dev.121145. PMID: 26534985
- Futura Gattobigio (en cours) – Université Paris Cité: Mechanism and role of NODAL signalling at preimplantation stages
- Giulia Tomaino (2024) – University of Milano-Bicocca (cotutelle/Dept of Biotechnology and Biosciences): Leveraging recombinant Vault nanoparticles produced in Komagataella phaffii for targeted delivery of siRNAs as therapeutic molecules
- Gaël Simon (2023) – Université Paris Cité: In vitro study of BMP, WNT and NODAL signalling pathways during primitive streak formation and specification using mouse 2D Gastruloids
- Nicolas Porchet (2019) – Université de Paris: Role of signaling pathways in cell-fate specification in the early mouse embryo
- Mathieu Vieira (2018) – Université Paris Diderot: Régulation de l’expression du gène Nodal lors de la différenciation des cellules souches embryonnaires
- Marieke Cajal (2013) – Université Paris Diderot: Étude des étapes précoces de la formation et de la régionalisation du cerveau des vertébrés : Un rôle conservé des cellules de l’ectoderme non-neural chez la souris et le poulet
- Elsa Mazari (2013) – Université Paris Sud (cotutelle/Laboratoire de Photonique et de Nanostructures, LPN-CNRS, Marcoussis): Microsystèmes magnétiques et électriques pour la modification spatio-temporelle de voies de signalisation biologiques
- Vasily Gurchenkov (2009) – Université Pierre et Marie Curie: Functional study of the asymmetric specific element of the Nodal gene during early mouse development
- Céline Granier (2007) – Université Paris Diderot: Étude de la dynamique de la voie de signalisation NODAL chez l’embryon de souris aux stades péri-implantatoires
Bertrand Cosson, EDC, Paris, France
- Gabriella MINCHIOTTI, IGB, Naples, Italie
- Claire Chazaud, Institut GReD, Clermont-Ferrand, France
- Jean-Léon MAITRE, Institut Curie, Paris, France
- Caroline HILL, Crick Institute, Londres, Grande-Bretagne
- Juliette AZIMZADEH, Institut Jacques Monod, Paris, France
- Benoit SORRE, Institut Curie, Paris, France
- Hervé ISAMBERT, Institut Curie, Paris, France
- Sandrine CABURET, Institut Jacques Monod, Paris, France
Former collaborations:
- Frédérique DESHAYES, Institut Jacques Monod, Paris, France
- Farida DJOUAD, IRMB, Montpellier, France
- Shoumo BHATTACHARYA, Radcliffe Dept of Medicine, University of Oxford, Oxford, UK
- Renata KOZYRAKI, Centre de Recherche des Cordeliers, Paris, France
- Michel COHEN-TANNOUDJI, Institut Pasteur, Paris, France
- Sophie CREUZET, Institut des Neurosciences Paris-Saclay, Université Paris-Saclay, Saclay, France
- An ZWIJSEN, Dept for Human Genetics, KU Leuven, Leuven, Belgium
- Susana M. CHUVA DE SOUSA LOPES, Leiden University Medical Centre, Leiden, The Netherlands
- Sascha OTT, Warwick Systems Biology Centre, University of Warwick, Coventry, UK
- Kirstie A. LAWSON, MRC Human Genetics Unit, University of Edinburgh, Edinburgh, UK
- Georgy KOENTGES, Wolfson Institute for Biomedical Research, University College London, London, UK
- Arnaud DUBOIS, Laboratoire d’Optique Physique, ESPCI, Paris, France
- Sylvie MAZAN, Développement et Évolution des Vertébrés, Université Paris-Sud, Orsay, France
- Bénédicte DURAND, CGMC, Université Claude Bernard Lyon-1, Villeurbanne
- Christian CIBERT, Institut Jacques Monod, Paris, France
Current sponsors:

Previous sponsors:



