Mitochondries, métaux et stress oxydatif
Jean-Michel CAMADRO
 L’étude des voies d’assimilation du fer et de son métabolisme intracellulaire chez un organisme modèle, la levure Saccharomyces cerevisiae, permet d’aborder des problèmes de biologie fondamentaux tout en envisageant des applications de ces recherches dans les domaines de la thérapeutique ou de la compréhension des bases moléculaires de pathologies humaines.
L’étude des voies d’assimilation du fer et de son métabolisme intracellulaire chez un organisme modèle, la levure Saccharomyces cerevisiae, permet d’aborder des problèmes de biologie fondamentaux tout en envisageant des applications de ces recherches dans les domaines de la thérapeutique ou de la compréhension des bases moléculaires de pathologies humaines.
Le fer est un oligo-élément essentiel pour toutes les cellules vivantes, mais l’excès ou le défaut d’apport en fer sont tous deux nocifs pour la cellule. L’homéostasie du fer est donc étroitement régulée dans la plupart, sinon la totalité, des systèmes vivants. Chez l’homme, les maladies liées à des anomalies de l’homéostasie du fer peuvent résulter soit de facteurs environnementaux, soit de certaines anomalies génétiques.
Le fer est le centre redox des hèmes et des centres fer-soufre, cofacteurs essentiels dans de nombreuses fonctions cellulaires, dont la production d’énergie et le métabolisme de l’oxygène. Un apport approprié en fer est donc obligatoire pour assurer une croissance et une survie normales des cellules. Cependant, en raison de sa réactivité chimique, le fer peut être toxique pour la cellule, et paradoxalement, la carence en fer et la surcharge en fer ont été associées à une production accrue d’espèces réactives de l’oxygène nuisibles (ROS).
Nous étudions plusieurs aspects du métabolisme du fer : son transport à travers la membrane plasmique et son utilisation dans les mitochondries, le contrôle de l’expression des gènes codant les composants des machines du métabolisme du fer dans différents systèmes modèles eucaryotes. Cela nous conduit à étudier en parallèle le contrôle des réponses thiol-dépendantes au stress oxydatif, une condition souvent associée aux altérations de l’homéostasie du fer.
Nos objets d’étude sont :
– des cellules humaines, pour mieux comprendre les bases moléculaires d’une maladie neurodégénérative fatale, l’ataxie de Friedreich
– des levures, S. cerevisiae comme système modèle, et C. albicans pour mieux comprendre le rôle du fer comme facteur de virulence
– des micro algues marines pour mieux comprendre les mécanismes d’adaptation du phytoplancton aux changements environnementaux majeurs : hausse de la température, augmentation du CO2, changement du pH du milieu marin.
Nous développons une méthode originale et très innovante de marquage métabolique (SLIM-Labeling) des différents types cellulaires que nous étudions pour analyser les variations quantitative d’abondance des protéines en réponse à différent stress biologiques. Nous appliquons en particulier cette nouvelle approcha à l’étude des bases moléculaires de la transition levure/hyphe et au contrôle épigénétique de cette transition.
Les approches de biologie computationnelles sont indispensables pour l’étude des jeux de données de grande dimension que nous produisons (transcriptomique, protéomique quantitative, phénomique), et nous contribuons activement à la production de nouveaux outils bioinformatiques « open source » et donc utilisables par la communauté scientifique.
Projet de recherche #1 (Valérie Serre)
L’ataxie de Friedreich (AF) est une maladie neurodégénérative héréditaire, due à une répétition GAA instable située dans l’intron 1 du gène FXN (9q21.11) codant pour la frataxine. Cette protéine liant le fer joue un rôle dans la biogenèse de clusters fer-soufre et le transport du fer dans la mitochondrie. Un déficit en cette protéine mitochondriale conduit à la perturbation progressive des systèmes nerveux central et périphérique observée dans l’AF. La taille de l’allèle le plus court est inversement proportionnelle à l’âge d’apparition de la maladie et du temps entre l’apparition des signes cliniques et le confinement en fauteuil roulant. Elle est corrélée de manière positive à la prévalence de la cardiomyopathie.
Depuis de nombreuses années de gros efforts ont été faits pour élaborer un traitement visant à améliorer les symptômes cliniques de l’AF. Toutefois, la mise au point d’un traitement efficace demeure un défi car la physiopathologie de l’AF demeure non totalement résolue. La découverte de biomarqueurs dans les maladies génétiques rares permet d’accélérer la recherche médicale en mettent en lumière les mécanismes physiopathologiques des maladies. Mis à part la mesure de l’expression de la frataxine, il n’existe pas à l’heure actuelle de biomarqueurs protéiques robustes connus pour être liés à la gravité ou à la progression de l’AF. De plus, les connaissances actuelles sur l’AF offrent diverses approches thérapeutiques à l’étude, mais il n’existe aucun biomarqueur fiable pour évaluer l’efficacité des traitements.
Notre projet vise à accélérer la découverte de cibles thérapeutiques par une approche de protéomique quantitative à haut débit, le but étant de pouvoir mettre en évidence des biomarqueurs d’intérêt à partir d’échantillons de plasmas de patients AF.
Les données brutes issues de la protéomique quantitative sont analysées en utilisant des outils statistiques et bioinformatiques selon un pipeline que nous avons mis au point. Ce pipeline d’analyse utilise des scripts en langage R inclus dans un fichier R Markdown, permettant de générer des rapports d’analyse détaillés, commentés, et reproductibles, ainsi que l’utilisation de plateformes logicielles en open source. Toutes ces analyses bioinformatiques suivent les principes généraux de science reproductible avec FAIR (Findable, Accessible, Interoperable and Reproducible). La validation de ces biomarqueurs potentiels pourra être facilement effectuée par Western blot ou tests ELISA en routine.
Ces travaux, effectués en étroite collaboration avec la plateforme de protéomique de l’Institut Jacques Monod et l’Institut du Cerveau (Paris), conduiront, nous l’espérons, à l’identification de marqueurs protéiques spécifiques du suivi des patients AF et à la mise en place de stratégies thérapeutiques ciblées et efficaces.
Projet de recherche #2 (Françoise Auchère)
Régulation de l’état “redox” cellulaire et réponse adaptative du pathogène opportuniste Candida albicans.
Candida albicans est un champignon normalement présent au niveau des cavités orales, de la peau et des muqueuses vaginales et digestives. C’est une levure dimorphique que l’on peut trouver sous forme de levure ou filamentée et la capacité d’adaptation de ce pathogène opportuniste aux différentes conditions environnementales joue un rôle important dans le phénomène de virulence.
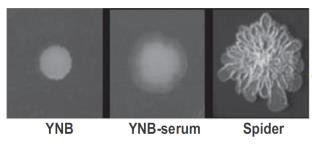
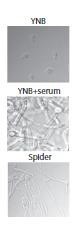 Nous nous intéressons à la réponse métabolique adaptative de C. albicans en présence de différentes sources de carbone à la fois sur les formes levure et les formes filamentées. Par des approches combinées de biochimie et de microbiologie, ainsi que par des approches de protéomique globale, nous étudions les différentes adaptations métaboliques associées au processus de filamentation, et les modifications de la fonction mitochondriale qui pourraient permettre une adaptation au cours de l’invasion des tissus hôtes.
Nous nous intéressons à la réponse métabolique adaptative de C. albicans en présence de différentes sources de carbone à la fois sur les formes levure et les formes filamentées. Par des approches combinées de biochimie et de microbiologie, ainsi que par des approches de protéomique globale, nous étudions les différentes adaptations métaboliques associées au processus de filamentation, et les modifications de la fonction mitochondriale qui pourraient permettre une adaptation au cours de l’invasion des tissus hôtes.
Projet de recherche #3 (Emmanuel Lesuisse, décédé en Août 2021 ; projet continué en collaboration avec Robert Sutack, Biocev Prague)
La biodisponibilité des métaux, et du fer en particulier est très impactée par la modification des écosystèmes marins due aux activités humaines (augmentation du taux de CO2 et acidification des océans). De manière surprenante, les métallomes/métalloprotéomes du phytoplancton sont peu caractérisés, et les prédictions relatives à l’évolution de la biodisponibilité du fer sont contradictoires. Notre but est de remédier à ce manque de données et de modéliser les effets de l’acidification des océans sur le cycle biogéochimique du fer. Une importante réserve de fer est associée à la surface de différentes espèces de phytoplancton. Nous étudions ce pool de fer en fonction de la pCO2 par des techniques biochimiques et biophysiques (études de sorption et techniques basées sur le synchrotron). Nous caractérisons les métallomes d’espèces cultivées sous différentes pCO2 en présence de différents métaux et nous étudions les effets des métaux et de la pCO2 sur la compétition interspécifique. La réponse au pH est aussi étudiée chez l’algue modèle Ostreococcus tauri.
Projet de recherche #4 (Jean-Michel Camadro)
Nouvelle méthode de marquage métabolique pour la quantification des variations d’abondance des protéines (intactes), à l’échelle des protéomes.
Nos approches expérimentales font largement appel aux analyses protéomiques quantitatives de type « label free ». En collaboration avec les ingénieurs de la plateforme protéomique de l’IJM, nous avons développé une nouvelle méthode d’analyse, le « SLIM-labeling » qui utilise la capacité qu’ont la plupart des systèmes vivants à synthétiser les acides aminés non-essentiels constitutifs des protéines à partir d’une source de carbone unique, telle que le glucose.
Le SLIM-Labeling vise à augmenter le taux de l’isotope léger (12C) du cardone dans les protéines, ce qui améliore de façon très significative la précision de la mesure de la masse des protéines, et permet le développement d’approches quantitatives originales.
Nous appliquons cette méthode à l’étude de l’abondance des protéines ribosomales chez la levure S. cerevisiae, à l’étude de la transition levure/hyphe chez C. albicans, et son contrôle épigénétique, au développement du nématode C. elegans (collaboration Equipe Dumont, IJM) et au marquage de cellules humaines en culture (collaboration Equipe Ladoux/ Mège, IJM)
Projet de recherche transversal (Pierre Poulain)
Développement des méthodes et des outils pour l’exploration, l’analyse et la visualisation de données biologiques, notamment en protéomique.
Parmi ces méthodes et outils développés dans le groupe, on peut citer :
– La bibliothèque Python AutoClassWrapper et le service web AutoClassWeb pour la classification non-supervisée Bayesienne de résultats expérimentaux omiques.
– La méthode bSLIM pour l’analyse quantitative en spectrométrie de masse par marquage métabolique avec un isotope léger.
– Enfin, le projet collaboratif Minomics a pour objectif l’analyse multi-omique de réseaux biologiques par une approche holistique.
Pierre est également ambassadeur Software Heritage afin promouvoir l’archivage des logiciels scientifiques open sources.
Robert SUTAK (Biocev, Prague, visiting scientist)
Alexia LOURENÇO (Visiting scientist)
Ancien membre :
Emmanuel LESUISSE ( Directeur de recherche)
Pour contacter un membre de l’équipe par mail : prenom.nom@ijm.fr
Denecker T, Zhou Li Y, Fairhead C, Budin K, Camadro JM, Bolotin-Fukuhara M, Angoulvant A, Lelandais G.
NAR Genom Bioinform. 2020 Apr 20;2(2):lqaa027. doi: 10.1093/nargab/lqaa027.
PMID: 33575583; PMCID: PMC7671338.
Sénécaut N, Alves G, Weisser H, Lignières L, Terrier S, Yang-Crosson L, Poulain P, Lelandais G, Yu YK, Camadro JM.
J Proteome Res. 2021 Mar 5;20(3):1476-1487.
doi: 10.1021/acs.jproteome.0c00478. Epub 2021 Feb 11.
PMID: 33573382; PMCID: PMC8459934.
3: Iron Uptake Mechanisms in Marine Phytoplankton.
Sutak R, Camadro JM, Lesuisse E.
Front Microbiol. 2020 Nov 5;11:566691. doi:10.3389/fmicb.2020.566691.
PMID: 33250865; PMCID: PMC7676907.
Duval C, Macabiou C, Garcia C, Lesuisse E, Camadro JM, Auchère F.
Microbiologyopen. 2020 Feb;9(2):e970. doi:10.1002/mbo3.970. Epub 2019 Dec 1.
PMID: 31788966; PMCID: PMC7002100.
El Banna N, Hatem E, Heneman-Masurel A, Léger T, Baïlle D, Vernis L, Garcia C, Martineau S, Dupuy C, Vagner S, Camadro JM, Huang ME.
Redox Biol. 2019 Sep;26:101290. doi: 10.1016/j.redox.2019.101290. Epub 2019 Aug 2.
PMID: 31412312; PMCID: PMC6831881.
6: NDUFS6 related Leigh syndrome: a case report and review of the literature.
Rouzier C, Chaussenot A, Fragaki K, Serre V, Ait-El-Mkadem S, Richelme C, Paquis-Flucklinger V, Bannwarth S.
J Hum Genet. 2019 Jul;64(7):637-645. doi: 10.1038/s10038-019-0594-4. Epub 2019 Apr 4.
PMID: 30948790.
Bossenmeyer-Pourié C, Smith AD, Lehmann S, Deramecourt V, Sablonnière B, Camadro JM, Pourié G, Kerek R, Helle D, Umoret R, Guéant-Rodriguez RM, Rigau V, Gabelle A, Sequeira JM, Quadros EV, Daval JL, Guéant JL.
J Pathol. 2019 Jul;248(3):291-303. doi: 10.1002/path.5254. Epub 2019 Mar 19.
PMID: 30734924.
Télot L, Rousseau E, Lesuisse E, Garcia C, Morlet B, Léger T, Camadro JM, Serre V.
Biochim Biophys Acta Mol Basis Dis. 2018 Apr;1864(4 Pt
A):997-1009. doi: 10.1016/j.bbadis.2018.01.010. Epub 2018 Jan 9.
PMID: 29329987.
Publications
Nouvelles approches de quantification des variations du protéome au niveau des protéines intactes : analyses expérimentales et computationnelles
Katerina Zeniskova (Robert Sutak, DT, Jean-Michel Camadro external supervisor)
Effect of various stress factors on mitochondrial processes of pathogenic protists
Thibaut Poinsignon (Gaëlle lLelandais, DT, Pierre Poulain, co-DT)
Modélisation des réseaux biologiques, analyse de données multi-omiques, visualisation à large échelle
Pr Gaëlle Lelandais, I2BC, Université Paris Saclay
Dr Yi-Kuo Yu, NCBI, NIH, Bethesda, USA
Pr Denis Mestivier, UPEC
Dr Stéphane Lemaire, IBPS, Sorbonne Université
Pr Jean-Louis Guéant, INSERM, Université de Lorraine
Dr Robert Sutac, Biocev, Prague, Rep. Tchèque
Pr Pavel Martasek, 1st Faculty of Medicine, Charles University, Prague, Rep. Tchèque
Data mining, Visualization and Computational Modeling of Redox Signaling Networks – MinOmics
2020-03 to 2024-03 | Grant
National Agency for Research (Paris, FR)
URL: https://app.dimensions.ai/details/grant/grant.9466085
GRANT_NUMBER: ANR-19-CE45-0017
Microbial Communities in Biomedical and Environmental Areas, and Systems Biology
2018-09 to 2022-11 | Grant
European Commission (Brussels, BE)
URL: https://app.dimensions.ai/details/grant/grant.7819194
GRANT_NUMBER: 810224
High performance MS-based proteomics by reducing stable isotopes complexity in vivo – SLIM-labeling
2018-09 to 2022-03 | Grant
National Agency for Research (Paris, FR)
URL: https://app.dimensions.ai/details/grant/grant.8639705
GRANT_NUMBER: ANR-18-CE44-0014
Le protéome urinaire dans l’ataxie de Friedreich : apport de la protéomique quantitative pour la découverte de biomarqueurs 2019-2019-2022 | Grant
Association Française Ataxie de Friedreich (AFAF, Paris FR)
URL : www.afaf.asso.fr
29 novembre 2022 : Inauguration du nouveau spectromètre de masse

