Research
The main objective of the research carried out by our team is the study of ovarian function through molecular genetics. A historical subject of the group has been the analysis of the Blepharophimosis syndrome (craniofacial anomalies and premature ovarian insufficiency or POI), due to alterations in the transcription factor FOXL2. Over the years, we have contributed to a better understanding of the molecular function of FOXL2, its mechanisms of pathogenicity and the discovery of numerous targets and partners. FOXL2 provides an excellent model to study the combinatorial regulation of transcription, as it modulates a series of apparently unrelated processes (e.g. cholesterol and steroid metabolism, apoptosis, cell cycle, free radical detoxification, etc.). Thus, we use FOXL2 as a model to explore the basis of specific target recognition by a transcription factor and the influence of its partners and post-translational modifications on this process. To do so, we use state-of-the-art genomics and proteomics tools.
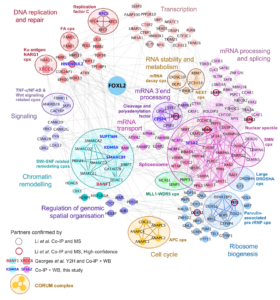
Caption: FOXL2 interactome: protein-protein interaction network for 255 FOXL2 partners identified by co-immunoprecipitation and mass spectrometry.
FOXL2 has also opened an avenue of research into ovarian granulosa cell tumors (GCTs), which account for up to 8% of all ovarian cancers. Two distinct subtypes of GCTs have been described: juvenile and adult forms. The adult type occurs most often during perimenopause and is characterized by late recurrence (up to 40 years after treatment of the primary tumor). The juvenile form (JGCT) tends to occur in the prepubertal period. A recurrent somatic mutation in FOXL2, leading to the p.C134W substitution has been detected in more than 97% of AGCTs, suggesting that this mutation is a driver of their formation. We are actively investigating the pathogenic mechanisms of this mutation using CRISPR-Cas9 modified cell lines, an animal model and genomic tools.
The molecular basis of JGCT is less well understood. We recently found tandem duplications in the reading frame of the AKT1 oncogene that affect the pleckstrin-homology domain (PHD) of the protein in over 60% of JGCTs. The effects of these mutations are currently being studied in cell and Drosophila models.
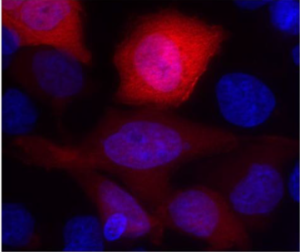
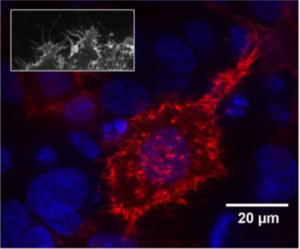
Caption: HeLa cells expressing the normal version of AKT1 (right) and one of the mutated versions found in ovarian tumors (left) (core: blue, AKT1: red).
Finally, we are taking advantage of the power of current genomic technology to discover new genes involved in female infertility due to POI, by performing exome sequencing in familial and isolated cases of this disorder. The functions of candidate genes discovered in these analyses are or will be studied using molecular approaches and animal models.
Members
Team leader
Reiner VEITIA,
Professor,
VEITIA LAB+33 (0)1 57 27 81 16, room 526B
Members
Sandrine CABURET,
Assistant Professor,
VEITIA LAB+33 (0)1 57 27 81 14, room 526B
Bérangère LEGOIS,
Biology technician,
VEITIA LAB+33 (0)1 57 27 80 81, room 526B
Ludovic MOUSSERON,
PhD student,
VEITIA LAB+33 (0)1 57 27 81 16, room 526B
Anne-Laure TODESCHINI,
Professor,
VEITIA LAB+33 (0)1 57 27 81 17, room 526B
Anis TOUSSIRT,
PhD student,
VEITIA LAB Alain ZIDER,
Professor,
VEITIA LAB+33 (0)1 57 27 80 85, room 526B
Despoina Maria CHOUSIANITI, Erasmus
To contact a member of the team by e-mail: name.surname@ijm.fr
Selected publications
Reply to “An alternative miRISC targets a cancer-associated coding sequence mutation in FOXL2”. Veitia RA, Pilsworth J, Todeschini AL, Huntsman D.EMBO J. 2021 Aug 16;40(16):e107517. doi: 10.15252/embj.2020107517.PMID: 34396573
FOXL2 in adult-type granulosa cell tumour of the ovary: oncogene or tumour suppressor gene? Pilsworth JA, Todeschini AL, Neilson SJ, Cochrane DR, Lai D, Anttonen M, Heikinheimo M, Huntsman DG, Veitia RA.J Pathol. 2021 Nov;255(3):225-231. doi: 10.1002/path.5771. Epub 2021 Sep 1.PMID: 34338304
Insights into the pathogenicity of missense variants in the forkhead domain of FOX proteins underlying Mendelian disorders. Bermúdez-Guzmán L, Veitia RA. Hum Genet. 2021 Jul;140(7):999-1010.
Forkhead Transcription Factors in Health and Disease. Herman L, Todeschini AL, Veitia RA.Trends Genet. 2021 May;37(5):460-475 (Review).
Genomic exploration of the targets of FOXL2 and ESR2 unveils their implication in cell migration, invasion, and adhesion. Herman L, Legois B, Todeschini AL, Veitia RA.FASEB J. 2021 Apr;35(4):e21355. doi: 10.1096/fj.202002444R.PMID: 33749886
A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1. Felipe-Medina N, Caburet S, Sánchez-Sáez F, Condezo YB, de Rooij DG, Gómez-H L, Garcia-Valiente R, Todeschini AL, Duque P, Sánchez-Martin MA, Shalev SA, Llano E, Veitia RA, Pendás AM.Elife. 2020 Aug 26;9:e56996. doi: 10.7554/eLife.56996.PMID: 32845237 Free
An exome-wide exploration of cases of primary ovarian insufficiency uncovers novel sequence variants and candidate genes. Alvarez-Mora MI, Todeschini AL, Caburet S, Perets LP, Mila M, Younis JS, Shalev S, Veitia RA.Clin Genet. 2020 Sep;98(3):293-298. doi: 10.1111/cge.13803. Epub 2020 Jul 28.PMID: 32613604
DHH pathogenic variants involved in 46,XY disorders of sex development differentially impact protein self-cleavage and structural conformation. Elzaiat M, Flatters D, Sierra-Díaz DC, Legois B, Laissue P, Veitia RA.Hum Genet. 2020 Nov;139(11):1455-1470.
Conventional and unconventional interactions of the transcription factor FOXL2 uncovered by a proteome-wide analysis. Penrad-Mobayed M, Perrin C, Herman L, Todeschini AL, Nigon F, Cosson B, Caburet S, Veitia RA.FASEB J. 2020 Jan;34(1):571-587. doi: 10.1096/fj.201901573R. Epub 2019 Nov 25.PMID: 31914586
A truncating MEIOB mutation responsible for familial primary ovarian insufficiency abolishes its interaction with its partner SPATA22 and their recruitment to DNA double-strand breaks. Caburet S, Todeschini AL, Petrillo C, Martini E, Farran ND, Legois B, Livera G, Younis JS, Shalev S, Veitia RA.EBioMedicine. 2019 Apr;42:524-531. doi: 10.1016/j.ebiom.2019.03.075. Epub 2019 Apr 15.PMID: 31000419
Publications
Publications
2913254
3ZUDK9KF
1
apa
50
date
desc
8863
https://www.ijm.fr/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A50%2C%22request_next%22%3A50%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22CRLZ49AC%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%20et%20al.%22%2C%22parsedDate%22%3A%222025-11-12%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%2C%20Herman%2C%20L.%2C%20Legois%2C%20B.%2C%20Claret%2C%20S.%2C%20Zider%2C%20A.%2C%20%26amp%3B%20Todeschini%2C%20A.-L.%20%282025%29.%20Deciphering%20the%20impact%20of%20AKT1%20pathogenic%20variants%20in%20Juvenile%20Granulosa%20Cell%20Tumors%20Using%20a%20Drosophila%20model.%20%26lt%3Bi%26gt%3BMolecular%20%26amp%3B%20Cellular%20Proteomics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B0%26lt%3B%5C%2Fi%26gt%3B%280%29.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.mcpro.2025.101466%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.mcpro.2025.101466%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Deciphering%20the%20impact%20of%20AKT1%20pathogenic%20variants%20in%20Juvenile%20Granulosa%20Cell%20Tumors%20Using%20a%20Drosophila%20model.%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laetitia%22%2C%22lastName%22%3A%22Herman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B%5Cu00e9rang%5Cu00e8re%22%2C%22lastName%22%3A%22Legois%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandra%22%2C%22lastName%22%3A%22Claret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alain%22%2C%22lastName%22%3A%22Zider%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%5D%2C%22abstractNote%22%3A%22%26lt%3Bh2%26gt%3BAbstract%26lt%3B%5C%2Fh2%26gt%3B%26lt%3Bh3%26gt%3BBackground%26lt%3B%5C%2Fh3%26gt%3B%26lt%3Bp%26gt%3BJuvenile-type%20granulosa%20cell%20tumors%20%28JGCTs%29%20manifest%20during%20the%20prepubertal%20period%20as%20precocious%20pseudo-puberty%20and%5C%2For%20dysmenorrhea.%20We%20have%20previously%20identified%20pathogenic%20variants%20in%20AKT1%20in%20JGCTs.%20This%20study%20aims%20to%20understand%20how%20these%20variants%20affect%20cellular%20function%20at%20the%20phenotypic%20and%20molecular%20levels%20using%20a%20Drosophila%20model.%26lt%3B%5C%2Fp%26gt%3B%26lt%3Bh3%26gt%3BMethods%26lt%3B%5C%2Fh3%26gt%3B%26lt%3Bp%26gt%3BTransgenic%20Drosophila%20models%20expressing%20wild-type%20%28WT%29%20AKT1%20and%20four%20pathogenic%20variants%20were%20created%20under%20the%20control%20of%20tissue-specific%20promoters.%20Phenotypic%20effects%20were%20studied%20by%20assessing%20%26lt%3Bi%26gt%3BDrosophila%26lt%3B%5C%2Fi%26gt%3B%20wings%20for%20cell%20division%20and%20growth%20using%20wing%20surface%20and%20trichome%20density%20and%20ovarian%20follicular%20cells%20were%20examined%20for%20subcellular%20localization%20and%20morphology.%20Molecular%20analyses%20included%20Mass%20Spectrometry%20%28MS%29%20to%20identify%20differentially%20expressed%20proteins%20%28DEPs%29%20and%20phospho-peptides%2C%20along%20with%20RNAseq%20to%20characterize%20transcriptomic%20changes.%26lt%3B%5C%2Fp%26gt%3B%26lt%3Bh3%26gt%3BResults%26lt%3B%5C%2Fh3%26gt%3B%26lt%3Bp%26gt%3BWings%20expressing%20mutated%20AKT1%20showed%20increased%20surface%20area%20and%20reduced%20trichome%20density%2C%20indicating%20larger%20cells.%20In%20ovarian%20follicular%20cells%2C%20WT%20AKT1%20localized%20primarily%20to%20the%20cytoplasm%2C%20while%20mutated%20AKT1%20variants%20were%20associated%20with%20the%20plasma%20membrane%2C%20leading%20to%20morphological%20abnormalities%20and%20increased%20cell%20size.%20MS%20revealed%20numerous%20DEPs%20and%20phospho-peptides%2C%20highlighting%20changes%20in%20pathways%20such%20as%20glycolysis%20and%20Rho%20GTPase%20signaling.%20Transcriptomics%20demonstrated%20a%20clear%20gain-of-function%20for%20mutated%20AKT1%20in%20activating%20a%20subset%20of%20genes.%20However%2C%20several%20genes%20up-regulated%20by%20WT%20AKT1%20were%20less%20effectively%20activated%20by%20the%20mutants%2C%20indicating%20a%20potential%20loss-of-function%20in%20transcriptional%20regulation%20for%20this%20subset%2C%20revealing%20an%20unexpected%20mechanistic%20complexity.%26lt%3B%5C%2Fp%26gt%3B%26lt%3Bh3%26gt%3BConclusions%26lt%3B%5C%2Fh3%26gt%3B%26lt%3Bp%26gt%3BNetwork%20analysis%20of%20interactions%20involving%20DEPs%2C%20phosphorylated%20proteins%2C%20and%20transcription%20factors%20suggests%20these%20elements%20mediate%20the%20observed%20proteomic%20and%20transcriptional%20alterations.%20Taken%20together%2C%20the%20results%20underscore%20the%20utility%20of%20Drosophila%20models%20in%20unraveling%20the%20biological%20relevance%20of%20AKT1%20pathogenic%20variants%20in%20cancer.%26lt%3B%5C%2Fp%26gt%3B%22%2C%22date%22%3A%222025%5C%2F11%5C%2F12%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.mcpro.2025.101466%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.mcponline.org%5C%2Farticle%5C%2FS1535-9476%2825%2900565-1%5C%2Fabstract%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221535-9476%2C%201535-9484%22%2C%22language%22%3A%22English%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222025-11-25T15%3A24%3A34Z%22%7D%7D%2C%7B%22key%22%3A%22V9WHF7XR%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%20et%20al.%22%2C%22parsedDate%22%3A%222025-04-17%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%2C%20Cowles%2C%20J.%20D.%2C%20%26amp%3B%20Caburet%2C%20S.%20%282025%29.%20Reclassifying%20NOBOX%20variants%20in%20primary%20ovarian%20insufficiency%20cases%20with%20a%20corrected%20gene%20model%20and%20a%20novel%20quantitative%20framework.%20%26lt%3Bi%26gt%3BHuman%20Reproduction%26lt%3B%5C%2Fi%26gt%3B%2C%20deaf058.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fhumrep%5C%2Fdeaf058%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fhumrep%5C%2Fdeaf058%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Reclassifying%20NOBOX%20variants%20in%20primary%20ovarian%20insufficiency%20cases%20with%20a%20corrected%20gene%20model%20and%20a%20novel%20quantitative%20framework%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jamie%20D%22%2C%22lastName%22%3A%22Cowles%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Caburet%22%7D%5D%2C%22abstractNote%22%3A%22How%20updated%20expression%20and%20genomic%20data%20combined%20with%20a%20disease%5C%2Fdisorder-specific%20classification%20system%20can%20be%20used%20to%20correct%20a%20gene%20model%20for%20a%20better%20evaluation%20of%20the%20pathogenicity%20of%20variants%20found%20in%20patients%3FBy%20combining%20available%20genomic%20and%20transcriptomic%20data%20from%20several%20species%20and%20a%20quantitative%20classification%20framework%20with%20primary%20ovarian%20insufficiency%20%28POI%29-adjusted%20parameters%2C%20we%20correct%20the%20human%20NOBOX%20%28newborn%20ovary%20homeobox%29%20gene%20model%20and%20provide%20a%20reclassification%20of%20variants%20previously%20reported%20in%20POI%20cases.The%20NOBOX%20gene%2C%20encoding%20a%20gonad-specific%20transcription%20factor%20with%20a%20crucial%20role%20in%20early%20folliculogenesis%20and%20considered%20a%20major%20gene%20involved%20in%20POI%2C%20is%20currently%20described%20as%20being%20expressed%20as%20four%20transcripts%2C%20the%20longest%20one%20considered%20canonical.%20All%20the%20variants%20identified%20in%20POI%20cases%20have%20been%20evaluated%20according%20to%20this%20canonical%20transcript%2C%20and%20the%20various%20functional%20tests%20have%20been%20performed%20using%20the%20corresponding%20predicted%20protein.We%20refined%20and%20corrected%20the%20NOBOX%20gene%20model%20using%20available%20genomic%20and%20RNAseq%20data%20in%20human%20and%2016%20other%20mammalian%20species.%20Expression%20data%20were%20selected%20for%20tissue%20specificity%2C%20strand%20specificity%2C%20and%20coverage.%20The%20analysis%20of%20RNAseq%20data%20from%20different%20ovarian%20fetal%20stages%20allows%20for%20a%20time-course%20description%20of%20NOBOX%20isoforms.%20Literature%20was%20scanned%20to%20retrieve%20NOBOX%20variants%20reported%20in%20POI%20cases%2C%20and%20NOBOX%20variants%20present%20in%20ClinVar%20and%20GnomAD%204%20databases%20were%20also%20retrieved.Strand-specific%20RNAseq%20data%20from%20human%20fetal%20ovaries%20and%20human%20adult%20testes%20were%20analysed%20to%20infer%20the%20correct%20human%20NOBOX%20gene%20isoforms.%20The%20conservation%20of%20the%20gene%20structure%20was%20verified%20by%20combining%20the%20aligned%20genomic%20sequences%20from%2017%20mammalian%20species%20covering%20a%20wide%20phylogenetic%20range%20and%20the%20relevant%20RNAseq%20data.%20As%20changing%20a%20gene%20model%20implies%20a%20reclassification%20of%20variants%2C%20we%20set%20up%20a%20quantitative%20framework%20with%20updated%20variant%20frequencies%20from%20GnomAD4%20and%20POI-adjusted%20parameters%20following%20the%20American%20College%20of%20Medical%20Genetics%20and%20Genomics%5C%2FAssociation%20for%20Molecular%20Pathology%20%28ACMG%5C%2FAMP%29%20guidelines.%20Using%20this%20framework%2C%20we%20reclassified%2044%20NOBOX%20variants%20reported%20in%20POI%20patients%20and%20families%2C%20117%20NOBOX%20variants%20reported%20in%20ClinVar%2C%20and%202613%20NOBOX%20variants%20present%20in%20GnomAD4.The%20corrected%20NOBOX%20gene%20model%20proposes%20the%20invalidation%20of%20two%20transcripts%2C%20including%20the%20canonical%20one.%20The%20two%20correct%20isoforms%20were%20present%20in%20fetal%20ovarian%20samples%2C%20and%20only%20one%20was%20detected%20in%20adult%20testes.%20Only%2014%20variants%20remained%20as%20possibly%20causative%20for%20POI.%20Furthermore%2C%20this%20re-evaluation%20strongly%20suggests%20that%20NOBOX%20biallelic%20variants%20are%20the%20most%20likely%20cause%20of%20POI.Large%20tables%20are%20provided%20as%20supplementary%20data%20sets%20on%20the%20Zenodo%20repository.The%20proposed%20gene%20model%20is%20robust%20but%20relies%20on%20available%20transcriptomic%20data%20covering%20a%20range%20of%20time%20points%20and%20tissues.%20Our%20scoring%20system%20was%20manually%20adjusted%20and%20other%20laboratories%20can%20implement%20it%20with%20different%20parameters.For%20the%20NOBOX%20variants%20that%20cannot%20be%20considered%20pathogenic%20or%20causative%20anymore%2C%20the%20genome%5C%2Fexome%20sequencing%20data%20of%20the%20corresponding%20patients%20should%20be%20reanalysed.%20Furthermore%2C%20the%20functional%20studies%20performed%20using%20the%20obsolete%20coding%20sequence%20should%20be%20reconsidered.%20The%20corrected%20gene%20model%20should%20be%20taken%20into%20account%20when%20evaluating%20novel%20NOBOX%20variants%20identified%20in%20POI%20patients.%20Our%20results%20highlight%20the%20importance%20of%20the%20careful%20assessment%20of%20the%20most%20updated%20expression%20data%20for%20validating%20a%20gene%20model%2C%20enabling%20a%20correct%20evaluation%20of%20the%20pathogenicity%20of%20variants%20found%20in%20patients.%20The%20proposed%20quantitative%20framework%20developed%20here%20can%20be%20used%20for%20the%20classification%20of%20variants%20in%20other%20genes%20underlying%20POI.%20Furthermore%2C%20the%20global%20approach%20based%20on%20quantitatively%20adjusting%20the%20ACMG%5C%2FAMP%20guidelines%20could%20be%20extended%20to%20other%20inherited%20pathologies.This%20project%20was%20not%20funded.%20All%20the%20authors%20have%20no%20conflict%20of%20interest%20to%20disclose.%22%2C%22date%22%3A%222025-04-17%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fhumrep%5C%2Fdeaf058%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fhumrep%5C%2Fdeaf058%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221460-2350%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222025-04-28T08%3A32%3A58Z%22%7D%7D%2C%7B%22key%22%3A%228FH35FAL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222025-03-05%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282025%29.%20Rethinking%20transcription%20factor%20dynamics%20and%20transcription%20regulation%20in%20eukaryotes.%20%26lt%3Bi%26gt%3BTrends%20in%20Biochemical%20Sciences%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tibs.2025.01.012%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tibs.2025.01.012%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Rethinking%20transcription%20factor%20dynamics%20and%20transcription%20regulation%20in%20eukaryotes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Transcription%20factors%20%28TFs%29%20control%20gene%20expression%20by%20binding%20to%20specific%20DNA%20motifs%20in%20cis-regulatory%20elements.%20Cooperativity%20has%20been%20thought%20to%20ensure%20TF%20binding%20specificity.%20Recent%20research%20suggests%20that%2C%20at%20least%20in%20yeast%2C%20the%20role%20of%20cooperativity%20has%20probably%20been%20overemphasized.%20Consequently%2C%20synergy%20%5Cu2013%20the%20collective%20recruitment%20of%20the%20transcriptional%20machinery%20by%20TFs%20bound%20at%20multiple%20DNA%20sites%20%5Cu2013%20emerges%20as%20a%20more%20significant%20mechanism%20for%20achieving%20the%20specificity%20of%20the%20transcriptional%20response.%20Furthermore%2C%20I%20argue%20that%20the%20concentration%20of%20TFs%20within%20phase-separated%20nuclear%20condensates%20and%20their%20covalent%20modifications%20play%20an%20underappreciated%20but%20crucial%20role%20in%20sharpening%20transcriptional%20responses%20through%20complementary%20mechanisms.%20A%20model%20integrating%20cooperativity%2C%20synergy%2C%20post-translational%20modifications%2C%20and%20phase%20separation%20provides%20a%20comprehensive%20framework%20to%20explain%20dynamic%2C%20context-specific%20transcriptional%20responses%20in%20eukaryotes.%22%2C%22date%22%3A%222025-03-05%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tibs.2025.01.012%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.sciencedirect.com%5C%2Fscience%5C%2Farticle%5C%2Fpii%5C%2FS0968000425000271%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220968-0004%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222025-03-12T08%3A53%3A25Z%22%7D%7D%2C%7B%22key%22%3A%22P8YH54IT%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%20et%20al.%22%2C%22parsedDate%22%3A%222025%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%2C%20Zschocke%2C%20J.%2C%20%26amp%3B%20Birchler%2C%20J.%20A.%20%282025%29.%20Gene%20Dosage%20Sensitivity%20and%20Human%20Genetic%20Diseases.%20%26lt%3Bi%26gt%3BJournal%20of%20Inherited%20Metabolic%20Disease%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B48%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20e70058.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1002%5C%2Fjimd.70058%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1002%5C%2Fjimd.70058%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Gene%20Dosage%20Sensitivity%20and%20Human%20Genetic%20Diseases%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Johannes%22%2C%22lastName%22%3A%22Zschocke%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%20A.%22%2C%22lastName%22%3A%22Birchler%22%7D%5D%2C%22abstractNote%22%3A%22Here%20we%20review%20the%20historical%20background%20and%20contemporary%20insights%20into%20genetic%20dominance%2C%20focusing%20on%20haploinsufficiency%20%28HI%29%2C%20that%20is%2C%20when%20the%20function%20of%20only%20one%20allele%20of%20a%20gene%20is%20not%20enough%20to%20ensure%20a%20normal%20phenotype%20in%20a%20diploid%20organism.%20A%20related%20phenomenon%20is%20triplosensitivity%2C%20that%20is%2C%20pathogenic%20effects%20when%20there%20are%20three%20instead%20of%20two%20copies%20of%20some%20%26%23039%3Bgenes%26%23039%3B.%20The%20importance%20of%20gene%20dosage%20issues%20was%20realized%20in%20humans%20when%20whole%20chromosomal%20abnormalities%20%28aneuploidy%29%20could%20be%20linked%20to%20clinical%20phenotypes%20such%20as%20Down%2C%20Edwards%2C%20and%20Patau%20syndromes.%20Subsequently%2C%20subtler%20chromosomal%20deletions%20and%20duplications%20have%20been%20shown%20to%20be%20responsible%20for%20many%20developmental%20syndromes.%20In%20several%20cases%2C%20a%20dosage-sensitive%20gene%20mapping%20to%20the%20relevant%20regions%20has%20been%20implicated%20as%20causal.%20We%20delve%20into%20the%20mechanisms%20of%20HI%2C%20especially%20due%20to%20direct%20protein%20insufficiency%20and%20subunit%20imbalances%20in%20the%20context%20of%20multi-subunit%20complexes.%20We%20show%20how%20the%20nonlinearity%20inherent%20to%20the%20relationship%20between%20genotype%20and%20phenotype%20is%20responsible%20for%20the%20dominance%20of%20the%20underlying%20genetic%20variants.%20We%20also%20explore%20why%20increased%20gene%20dosage%20can%20lead%20to%20abnormal%20phenotypes.%20Examples%20include%20trisomy%20or%20segmental%20genomic%20duplications%20in%20humans%20and%20oncogene%20amplification%20in%20cancers.%20Finally%2C%20we%20examine%20a%20few%20cases%20of%20genetic%20synergy%2C%20where%20the%20combined%20effect%20of%20two%20or%20more%20variants%20amplifies%20their%20individual%20effects%2C%20underlying%20a%20distinguishable%20phenotype.%20Further%20research%20is%20required%20to%20elucidate%20the%20dynamics%20of%20multicomponent%20interactions%20to%20unravel%20the%20mechanistic%20complexities%20of%20genetic%20dominance%2C%20inter-gene%20interactions%2C%20and%20their%20implications%20for%20disease.%22%2C%22date%22%3A%222025%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1002%5C%2Fjimd.70058%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fonlinelibrary.wiley.com%5C%2Fdoi%5C%2Fabs%5C%2F10.1002%5C%2Fjimd.70058%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221573-2665%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222025-07-15T12%3A57%3A18Z%22%7D%7D%2C%7B%22key%22%3A%22BFLZAES2%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222024-10-26%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282024%29.%20Emerging%20links%20between%20phase%20separation%20and%20transcription%20factor%20haploinsufficiency.%20%26lt%3Bi%26gt%3BTrends%20in%20Genetics%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2024.10.001%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2024.10.001%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Emerging%20links%20between%20phase%20separation%20and%20transcription%20factor%20haploinsufficiency%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Recent%20studies%20have%20addressed%20the%20relevance%20of%20phase%20separation%2C%20by%20which%20membrane-less%20compartments%20are%20formed%20within%20the%20nucleus%2C%20to%20understand%20the%20impact%20of%20genetic%20variants.%20They%20highlight%20unsuspected%20links%20between%20phase%20separation%20and%20haploinsufficiency%20of%20transcription%20factors.%22%2C%22date%22%3A%222024-10-26%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tig.2024.10.001%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.sciencedirect.com%5C%2Fscience%5C%2Farticle%5C%2Fpii%5C%2FS016895252400235X%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220168-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222024-10-28T09%3A14%3A22Z%22%7D%7D%2C%7B%22key%22%3A%22QQPHTACW%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Herman%20et%20al.%22%2C%22parsedDate%22%3A%222024-03-01%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BHerman%2C%20L.%2C%20Amo%2C%20A.%2C%20Legois%2C%20B.%2C%20Di%20Carlo%2C%20C.%2C%20Veitia%2C%20R.%20A.%2C%20%26amp%3B%20Todeschini%2C%20A.-L.%20%282024%29.%20A%20cellular%20model%20provides%20insights%20into%20the%20pathogenicity%20of%20the%20oncogenic%20FOXL2%20somatic%20variant%20p.Cys134Trp.%20%26lt%3Bi%26gt%3BBritish%20Journal%20of%20Cancer%26lt%3B%5C%2Fi%26gt%3B%2C%201%26%23×2013%3B10.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41416-024-02613-x%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41416-024-02613-x%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20cellular%20model%20provides%20insights%20into%20the%20pathogenicity%20of%20the%20oncogenic%20FOXL2%20somatic%20variant%20p.Cys134Trp%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laetitia%22%2C%22lastName%22%3A%22Herman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ang%5Cu00e9lique%22%2C%22lastName%22%3A%22Amo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Berang%5Cu00e8re%22%2C%22lastName%22%3A%22Legois%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Caterina%22%2C%22lastName%22%3A%22Di%20Carlo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%5D%2C%22abstractNote%22%3A%22FOXL2%20is%20a%20transcription%20factor%20expressed%20in%20ovarian%20granulosa%20cells.%20A%20somatic%20variant%20of%20FOXL2%20%28c.402%5Cu2009C%5Cu2009%26gt%3B%5Cu2009G%2C%20p.Cys134Trp%29%20is%20the%20hallmark%20of%20adult-type%20granulosa%20cell%20tumours.%22%2C%22date%22%3A%222024-03-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41416-024-02613-x%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nature.com%5C%2Farticles%5C%2Fs41416-024-02613-x%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221532-1827%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222024-03-04T11%3A15%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22ZB43AKSH%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Zutterling%20et%20al.%22%2C%22parsedDate%22%3A%222023-11-02%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BZutterling%2C%20C.%2C%20Todeschini%2C%20A.-L.%2C%20Fourmy%2C%20D.%2C%20Busso%2C%20D.%2C%20Veaute%2C%20X.%2C%20Ducong%26%23xE9%3B%2C%20F.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282023%29.%20The%20forkhead%20DNA-binding%20domain%20binds%20specific%20G2-rich%20RNA%20sequences.%20%26lt%3Bi%26gt%3BNucleic%20Acids%20Research%26lt%3B%5C%2Fi%26gt%3B%2C%20gkad994.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkad994%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkad994%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20forkhead%20DNA-binding%20domain%20binds%20specific%20G2-rich%20RNA%20sequences%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Caroline%22%2C%22lastName%22%3A%22Zutterling%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Deborah%22%2C%22lastName%22%3A%22Fourmy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Didier%22%2C%22lastName%22%3A%22Busso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xavier%22%2C%22lastName%22%3A%22Veaute%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Ducong%5Cu00e9%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Transcription%20factors%20contain%20a%20DNA-binding%20domain%20ensuring%20specific%20recognition%20of%20DNA%20target%20sequences.%20The%20family%20of%20forkhead%20%28FOX%29%20transcription%20factors%20is%20composed%20of%20dozens%20of%20paralogs%20in%20mammals.%20The%20forkhead%20domain%20%28FHD%29%20is%20a%20segment%20of%20about%20100%20amino%20acids%20that%20binds%20an%20A-rich%20DNA%20sequence.%20Using%20DNA%20and%20RNA%20PCR-SELEX%2C%20we%20show%20that%20recombinant%20FOXL2%20proteins%2C%20either%20wild-type%20or%20carrying%20the%20oncogenic%20variant%20C134W%2C%20recognize%20similar%20DNA-binding%20sites.%20This%20suggests%20that%20the%20oncogenic%20variant%20does%20not%20alter%20the%20intrinsic%20sequence-specificity%20of%20FOXL2.%20Most%20importantly%2C%20we%20show%20that%20FOXL2%20binds%20G2-rich%20RNA%20sequences%20whereas%20it%20virtually%20fails%20to%20bind%20similar%20sequences%20in%20DNA%20chemistry.%20Interestingly%2C%20a%20statistically%20significant%20subset%20of%20genes%20responding%20to%20the%20knock-down%20of%20FOXL2%5C%2FFoxl2%20harbor%20such%20G2-rich%20sequences%20and%20are%20involved%20in%20crucial%20signaling%20pathways%20and%20cellular%20processes.%20In%20addition%2C%20we%20show%20that%20FOXA1%2C%20FOXO3a%20and%20chimeric%20FOXL2%20proteins%20containing%20the%20FHD%20of%20the%20former%20are%20also%20able%20to%20interact%20with%20some%20of%20the%20preferred%20FOXL2-binding%20sequences.%20Our%20results%20point%20to%20an%20unexpected%20and%20novel%20characteristic%20of%20the%20forkhead%20domain%2C%20the%20biological%20relevance%20of%20which%20remains%20to%20be%20explored.%22%2C%22date%22%3A%222023-11-02%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkad994%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkad994%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220305-1048%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-11-09T15%3A36%3A51Z%22%7D%7D%2C%7B%22key%22%3A%22BHIP2VRL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Pozzi%20et%20al.%22%2C%22parsedDate%22%3A%222023-03-23%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPozzi%2C%20C.%2C%20Vanet%2C%20A.%2C%20Francesconi%2C%20V.%2C%20Tagliazucchi%2C%20L.%2C%20Tassone%2C%20G.%2C%20Venturelli%2C%20A.%2C%20Spyrakis%2C%20F.%2C%20Mazzorana%2C%20M.%2C%20Costi%2C%20M.%20P.%2C%20%26amp%3B%20Tonelli%2C%20M.%20%282023%29.%20Antitarget%2C%20Anti-SARS-CoV-2%20Leads%2C%20Drugs%2C%20and%20the%20Drug%20Discovery-Genetics%20Alliance%20Perspective.%20%26lt%3Bi%26gt%3BJournal%20of%20Medicinal%20Chemistry%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B66%26lt%3B%5C%2Fi%26gt%3B%286%29%2C%203664%26%23×2013%3B3702.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facs.jmedchem.2c01229%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1021%5C%2Facs.jmedchem.2c01229%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Antitarget%2C%20Anti-SARS-CoV-2%20Leads%2C%20Drugs%2C%20and%20the%20Drug%20Discovery-Genetics%20Alliance%20Perspective%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cecilia%22%2C%22lastName%22%3A%22Pozzi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne%22%2C%22lastName%22%3A%22Vanet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Valeria%22%2C%22lastName%22%3A%22Francesconi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lorenzo%22%2C%22lastName%22%3A%22Tagliazucchi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giusy%22%2C%22lastName%22%3A%22Tassone%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alberto%22%2C%22lastName%22%3A%22Venturelli%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Francesca%22%2C%22lastName%22%3A%22Spyrakis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marco%22%2C%22lastName%22%3A%22Mazzorana%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maria%20P.%22%2C%22lastName%22%3A%22Costi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michele%22%2C%22lastName%22%3A%22Tonelli%22%7D%5D%2C%22abstractNote%22%3A%22The%20most%20advanced%20antiviral%20molecules%20addressing%20major%20SARS-CoV-2%20targets%20%28Main%20protease%2C%20Spike%20protein%2C%20and%20RNA%20polymerase%29%2C%20compared%20with%20proteins%20of%20other%20human%20pathogenic%20coronaviruses%2C%20may%20have%20a%20short-lasting%20clinical%20efficacy.%20Accumulating%20knowledge%20on%20the%20mechanisms%20underlying%20the%20target%20structural%20basis%2C%20its%20mutational%20progression%2C%20and%20the%20related%20biological%20significance%20to%20virus%20replication%20allows%20envisaging%20the%20development%20of%20better-targeted%20therapies%20in%20the%20context%20of%20COVID-19%20epidemic%20and%20future%20coronavirus%20outbreaks.%20The%20identification%20of%20evolutionary%20patterns%20based%20solely%20on%20sequence%20information%20analysis%20for%20those%20targets%20can%20provide%20meaningful%20insights%20into%20the%20molecular%20basis%20of%20host-pathogen%20interactions%20and%20adaptation%2C%20leading%20to%20drug%20resistance%20phenomena.%20Herein%2C%20we%20will%20explore%20how%20the%20study%20of%20observed%20and%20predicted%20mutations%20may%20offer%20valuable%20suggestions%20for%20the%20application%20of%20the%20so-called%20%26quot%3Bsynthetic%20lethal%26quot%3B%20strategy%20to%20SARS-CoV-2%20Main%20protease%20and%20Spike%20protein.%20The%20synergy%20between%20genetics%20evidence%20and%20drug%20discovery%20may%20prioritize%20the%20development%20of%20novel%20long-lasting%20antiviral%20agents.%22%2C%22date%22%3A%222023-03-23%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1021%5C%2Facs.jmedchem.2c01229%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221520-4804%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-05-03T12%3A57%3A20Z%22%7D%7D%2C%7B%22key%22%3A%222MSZT6N6%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Ariste%20et%20al.%22%2C%22parsedDate%22%3A%222023-02%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BAriste%2C%20O.%2C%20de%20la%20Grange%2C%20P.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282023%29.%20Recurrent%20missense%20variants%20in%20clonal%20hematopoiesis-related%20genes%20present%20in%20the%20general%20population.%20%26lt%3Bi%26gt%3BClinical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B103%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20247%26%23×2013%3B251.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.14259%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.14259%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Recurrent%20missense%20variants%20in%20clonal%20hematopoiesis-related%20genes%20present%20in%20the%20general%20population%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Olivier%22%2C%22lastName%22%3A%22Ariste%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pierre%22%2C%22lastName%22%3A%22de%20la%20Grange%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Clonal%20hematopoiesis%20%28CH%29%20consists%20in%20an%20abnormal%20expansion%20of%20a%20hematopoietic%20stem%20cell%20bearing%20an%20advantageous%20somatic%20variant.%20A%20survey%20of%20known%20recurrent%20somatic%20missense%20variants%20in%20DNMT3A%2C%20SF3B1%2C%20SRSF2%2C%20and%20TP53%2C%20some%20of%20the%20most%20prominent%20genes%20underlying%20CH%20of%20indeterminate%20potential%20%28CHIP%29%2C%20in%20gnomAD%20noncancer%20database%20shows%20the%20presence%20of%2073%20variants.%20Many%20of%20them%20reach%20frequencies%20higher%20than%200.01%25%20in%20various%20populations%20and%2C%20in%20many%20cases%2C%20are%20enriched%20in%20specific%20populations.%20Consistent%20with%20a%20potential%20involvement%20in%20CHIP%2C%20we%20found%20that%20the%20age%20distribution%20of%20the%20carriers%20is%20shifted%20towards%20old%20ages.%20Moreover%2C%20the%20variant%20allele%20frequencies%20are%20on%20average%20lower%20than%2050%25%2C%20expected%20for%20germline%20heterozygous%20variants.%20The%20pervasive%20presence%20of%20some%20of%20such%20variants%20in%20blood%20DNA%20from%20elder%20individuals%20is%20compatible%20with%20CHIP%20of%20somatic%20origin.%20On%20practical%20grounds%2C%20CHIP%20can%20lead%20to%20misclassification%20of%20somatic%20variants%20in%20cancer-predisposition%20genes%20as%20inherited%2C%20which%20bear%20consequences%20for%20the%20affected%20individuals%20and%20their%20families.%22%2C%22date%22%3A%222023-02%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fcge.14259%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221399-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-05-03T12%3A50%3A47Z%22%7D%7D%2C%7B%22key%22%3A%229DX6ZJZN%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Llano%20et%20al.%22%2C%22parsedDate%22%3A%222023-01-18%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BLlano%2C%20E.%2C%20Todeschini%2C%20A.%20L.%2C%20Felipe-Medina%2C%20N.%2C%20Corte-Torres%2C%20M.%20D.%2C%20Condezo%2C%20Y.%20B.%2C%20Sanchez-Martin%2C%20M.%2C%20L%26%23xF3%3Bpez-Tamargo%2C%20S.%2C%20Astudillo%2C%20A.%2C%20Puente%2C%20X.%20S.%2C%20Pendas%2C%20A.%20M.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282023%29.%20The%20Oncogenic%20FOXL2%20C134W%20Mutation%20Is%20a%20Key%20Driver%20of%20Granulosa%20Cell%20Tumors.%20%26lt%3Bi%26gt%3BCancer%20Research%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B83%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20239%26%23×2013%3B250.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1158%5C%2F0008-5472.CAN-22-1880%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1158%5C%2F0008-5472.CAN-22-1880%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20Oncogenic%20FOXL2%20C134W%20Mutation%20Is%20a%20Key%20Driver%20of%20Granulosa%20Cell%20Tumors%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elena%22%2C%22lastName%22%3A%22Llano%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne%20Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Natalia%22%2C%22lastName%22%3A%22Felipe-Medina%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mar%5Cu00eda%20D.%22%2C%22lastName%22%3A%22Corte-Torres%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yazmine%20B.%22%2C%22lastName%22%3A%22Condezo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Manuel%22%2C%22lastName%22%3A%22Sanchez-Martin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sara%22%2C%22lastName%22%3A%22L%5Cu00f3pez-Tamargo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aurora%22%2C%22lastName%22%3A%22Astudillo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xose%20S.%22%2C%22lastName%22%3A%22Puente%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alberto%20M.%22%2C%22lastName%22%3A%22Pendas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Adult-type%20granulosa%20cell%20tumors%20%28AGCT%29%20are%20the%20most%20common%20type%20of%20malignant%20ovarian%20sex%20cord-stromal%20tumors.%20Most%20AGCTs%20carry%20the%20somatic%20variant%20c.402C%26gt%3BG%20%28p.C134W%29%20affecting%20the%20transcription%20factor%20FOXL2.%20Germline%20dominant%20variants%20in%20FOXL2%20are%20responsible%20for%20blepharophimosis%20syndrome%2C%20which%20is%20characterized%20by%20underdevelopment%20of%20the%20eyelid.%20In%20this%20work%2C%20we%20generated%20a%20mouse%20model%20harboring%20the%20C134W%20variant%20of%20FOXL2%20to%20evaluate%20in%20vivo%20the%20poorly%20understood%20oncogenic%20role%20of%20FOXL2.%20The%20mutation%20was%20dominant%20regarding%20eyelid%20hypoplasia%2C%20reminiscent%20of%20blepharophimosis%20syndrome.%20Interestingly%2C%20Foxl2%2B%5C%2FC134W%20female%20mice%20had%20reduced%20fertility%20and%20developed%20AGCTs%20through%20a%20progression%20from%20abnormal%20ovaries%20with%20aberrant%20granulosa%20cells%20to%20ovaries%20with%20stromal%20hyperplasia%20and%20atypia%20and%20on%20to%20tumors%20in%20adut%20mice.%20The%20genes%20dysregulated%20in%20mouse%20AGCTs%20exhibited%20the%20hallmarks%20of%20cancer%20and%20were%20consistent%20with%20a%20gain-of-function%20of%20the%20mutated%20allele%20affecting%20TGF%5Cu03b2%20signaling.%20A%20comparison%20of%20these%20data%20with%20previous%20results%20on%20human%20AGCTs%20indicated%20similar%20deregulated%20pathways.%20Finally%2C%20a%20mutational%20analysis%20of%20mouse%20AGCT%20transcriptomic%20data%20suggested%20the%20absence%20of%20additional%20driver%20mutations%20apart%20from%20FOXL2-C134W.%20These%20results%20provide%20a%20clear%20in%20vivo%20example%20in%20which%20a%20single%20mutational%20hit%20triggers%20tumor%20development%20associated%20with%20profound%20transcriptomic%20alterations.%5CnSIGNIFICANCE%3A%20A%20newly%20generated%20mouse%20model%20carrying%20a%20FOXL2%20mutation%20characteristic%20of%20adult-type%20granulosa%20cell%20tumors%20shows%20that%20FOXL2%20C134W%20shifts%20the%20transcriptome%20towards%20a%20signature%20of%20granulosa%20cell%20cancer%20and%20drives%20tumorigenesis.%22%2C%22date%22%3A%222023-01-18%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1158%5C%2F0008-5472.CAN-22-1880%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221538-7445%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-05-03T12%3A57%3A20Z%22%7D%7D%2C%7B%22key%22%3A%226984CIQ6%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222022-08%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282022%29.%20Who%20ever%20thought%20genetic%20mutations%20were%20random%3F%20%26lt%3Bi%26gt%3BTrends%20in%20Plant%20Science%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B27%26lt%3B%5C%2Fi%26gt%3B%288%29%2C%20733%26%23×2013%3B735.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tplants.2022.03.003%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tplants.2022.03.003%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Who%20ever%20thought%20genetic%20mutations%20were%20random%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22In%20a%20recent%20study%20of%20de%20novo%20mutations%20in%20arabidopsis%20%28Arabidopsis%20thaliana%29%2C%20Monroe%20et%20al.%20found%20a%20lower%20mutation%20frequency%20inside%20gene%20bodies%20and%20certain%20essential%20genes%2C%20shattering%20the%20long-standing%20idea%20that%20mutations%20are%20entirely%20random%20across%20the%20genome.%20Here%20I%20highlight%20the%20molecular%20nonrandomness%20of%20mutations%20and%20the%20type%20of%20gene%20affected.%22%2C%22date%22%3A%222022-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tplants.2022.03.003%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-4372%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A25%3A09Z%22%7D%7D%2C%7B%22key%22%3A%222JAEY6DF%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Franca%20et%20al.%22%2C%22parsedDate%22%3A%222022-05-27%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BFranca%2C%20M.%20M.%2C%20Condezo%2C%20Y.%20B.%2C%20Elzaiat%2C%20M.%2C%20Felipe-Medina%2C%20N.%2C%20S%26%23xE1%3Bnchez-S%26%23xE1%3Bez%2C%20F.%2C%20Mu%26%23xF1%3Boz%2C%20S.%2C%20Sainz-Urruela%2C%20R.%2C%20Mart%26%23xED%3Bn-Herv%26%23xE1%3Bs%2C%20M.%20R.%2C%20Garc%26%23xED%3Ba-Valiente%2C%20R.%2C%20S%26%23xE1%3Bnchez-Mart%26%23xED%3Bn%2C%20M.%20A.%2C%20Astudillo%2C%20A.%2C%20Mendez%2C%20J.%2C%20Llano%2C%20E.%2C%20Veitia%2C%20R.%20A.%2C%20Mendonca%2C%20B.%20B.%2C%20%26amp%3B%20Pend%26%23xE1%3Bs%2C%20A.%20M.%20%282022%29.%20A%20truncating%20variant%20of%20RAD51B%20associated%20with%20primary%20ovarian%20insufficiency%20provides%20insights%20into%20its%20meiotic%20and%20somatic%20functions.%20%26lt%3Bi%26gt%3BCell%20Death%20and%20Differentiation%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41418-022-01021-z%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41418-022-01021-z%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20truncating%20variant%20of%20RAD51B%20associated%20with%20primary%20ovarian%20insufficiency%20provides%20insights%20into%20its%20meiotic%20and%20somatic%20functions%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Monica%20M.%22%2C%22lastName%22%3A%22Franca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yazmine%20B.%22%2C%22lastName%22%3A%22Condezo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ma%5Cu00ebva%22%2C%22lastName%22%3A%22Elzaiat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Natalia%22%2C%22lastName%22%3A%22Felipe-Medina%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fernando%22%2C%22lastName%22%3A%22S%5Cu00e1nchez-S%5Cu00e1ez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sergio%22%2C%22lastName%22%3A%22Mu%5Cu00f1oz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Raquel%22%2C%22lastName%22%3A%22Sainz-Urruela%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%20Rosario%22%2C%22lastName%22%3A%22Mart%5Cu00edn-Herv%5Cu00e1s%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rodrigo%22%2C%22lastName%22%3A%22Garc%5Cu00eda-Valiente%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Manuel%20A.%22%2C%22lastName%22%3A%22S%5Cu00e1nchez-Mart%5Cu00edn%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aurora%22%2C%22lastName%22%3A%22Astudillo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juan%22%2C%22lastName%22%3A%22Mendez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elena%22%2C%22lastName%22%3A%22Llano%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Berenice%20B.%22%2C%22lastName%22%3A%22Mendonca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alberto%20M.%22%2C%22lastName%22%3A%22Pend%5Cu00e1s%22%7D%5D%2C%22abstractNote%22%3A%22Primary%20ovarian%20insufficiency%20%28POI%29%20causes%20female%20infertility%20by%20abolishing%20normal%20ovarian%20function.%20Although%20its%20genetic%20etiology%20has%20been%20extensively%20investigated%2C%20most%20POI%20cases%20remain%20unexplained.%20Using%20whole-exome%20sequencing%2C%20we%20identified%20a%20homozygous%20variant%20in%20RAD51B%20-%28c.92delT%29%20in%20two%20sisters%20with%20POI.%20In%20vitro%20studies%20revealed%20that%20this%20variant%20leads%20to%20translation%20reinitiation%20at%20methionine%2064.%20Here%2C%20we%20show%20that%20this%20is%20a%20pathogenic%20hypomorphic%20variant%20in%20a%20mouse%20model.%20Rad51bc.92delT%5C%2Fc.92delT%20mice%20exhibited%20meiotic%20DNA%20repair%20defects%20due%20to%20RAD51%20and%20HSF2BP%5C%2FBMRE1%20accumulation%20in%20the%20chromosome%20axes%20leading%20to%20a%20reduction%20in%20the%20number%20of%20crossovers.%20Interestingly%2C%20the%20interaction%20of%20RAD51B-c.92delT%20with%20RAD51C%20and%20with%20its%20newly%20identified%20interactors%20RAD51%20and%20HELQ%20was%20abrogated%20or%20diminished.%20Repair%20of%20mitomycin-C-induced%20chromosomal%20aberrations%20was%20impaired%20in%20RAD51B%5C%2FRad51b-c.92delT%20human%20and%20mouse%20somatic%20cells%20in%20vitro%20and%20in%20explanted%20mouse%20bone%20marrow%20cells.%20Accordingly%2C%20Rad51b-c.92delT%20variant%20reduced%20replication%20fork%20progression%20of%20patient-derived%20lymphoblastoid%20cell%20lines%20and%20pluripotent%20reprogramming%20efficiency%20of%20primary%20mouse%20embryonic%20fibroblasts.%20Finally%2C%20Rad51bc.92delT%5C%2Fc.92delT%20mice%20displayed%20increased%20incidence%20of%20pituitary%20gland%20hyperplasia.%20These%20results%20provide%20new%20mechanistic%20insights%20into%20the%20role%20of%20RAD51B%20not%20only%20in%20meiosis%20but%20in%20the%20maintenance%20of%20somatic%20genome%20stability.%22%2C%22date%22%3A%222022-05-27%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41418-022-01021-z%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221476-5403%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A25%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22Y2BBMMEQ%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Barakizou%20et%20al.%22%2C%22parsedDate%22%3A%222022-03-03%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBarakizou%2C%20H.%2C%20Souha%2C%20G.%2C%20Kamoun%2C%20T.%2C%20Mehdi%2C%20M.%2C%20Amary%2C%20F.%2C%20Huma%2C%20Z.%2C%20Todeschini%2C%20A.%2C%20Veitia%2C%20R.%2C%20%26amp%3B%20Donaldson%2C%20M.%20%282022%29.%20Precocious%20Pseudo-puberty%20in%20a%20Two-year-old%20Girl%2C%20Presenting%20with%20Bilateral%20Ovarian%20Enlargement%20and%20Progressing%20to%20Unilateral%20Juvenile%20Granulosa%20Cell%20Tumour.%20%26lt%3Bi%26gt%3BJournal%20of%20Clinical%20Research%20in%20Pediatric%20Endocrinology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B14%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%20107%26%23×2013%3B113.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.4274%5C%2Fjcrpe.galenos.2021.2021.0039%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.4274%5C%2Fjcrpe.galenos.2021.2021.0039%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Precocious%20Pseudo-puberty%20in%20a%20Two-year-old%20Girl%2C%20Presenting%20with%20Bilateral%20Ovarian%20Enlargement%20and%20Progressing%20to%20Unilateral%20Juvenile%20Granulosa%20Cell%20Tumour%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hager%22%2C%22lastName%22%3A%22Barakizou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gannouni%22%2C%22lastName%22%3A%22Souha%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thouraya%22%2C%22lastName%22%3A%22Kamoun%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Muhammed%22%2C%22lastName%22%3A%22Mehdi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fernanda%22%2C%22lastName%22%3A%22Amary%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Zilla%22%2C%22lastName%22%3A%22Huma%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Malcolm%22%2C%22lastName%22%3A%22Donaldson%22%7D%5D%2C%22abstractNote%22%3A%22Ovarian%20causes%20of%20precocious%20pseudo-puberty%20%28PPP%29%20include%20McCune-Albright%20syndrome%20%28MAS%29%20and%20juvenile%20granulosa%20cell%20tumour%20%28JGCT%29.%20We%20describe%20a%20case%20of%20PPP%20in%20which%20bilateral%20ovarian%20enlargement%20with%20multiple%20cysts%20progressed%20to%20unilateral%20JGCT.%20A%20girl%20aged%202.17%20years%20presented%20with%20three%20months%20of%20breast%20development%2C%20and%20rapid%20growth.%20Examination%20showed%20tall%20stature%2C%20height%20%2B2.6%20standard%20deviations%2C%20Tanner%20stage%20B3P2A1.%20A%20single%20caf%5Cu00e9%20au%20lait%20patch%20was%20noted.%20Bone%20age%20was%20advanced%20at%205%20years.%20Pelvic%20ultrasound%20showed%20bilaterally%20enlarged%20ovaries%20%28estimated%20volumes%2076%20mL%20on%20the%20left%2C%20139%20mL%20on%20the%20right%29%2C%20each%20containing%20multiple%20cysts.%20Luteinizing%20hormone%20%28LH%29%20and%20follicle%20stimulating%20hormone%20%28FSH%29%20values%20before%5C%2Fafter%20gonadotrophin%20administration%20were%200.43%5C%2F0.18%20and%20%26lt%3B0.1%5C%2F%26lt%3B0.1%20mUI%5C%2FmL%2C%20serum%20estradiol%20130%20pg%5C%2FmL%2C%20%28prepubertal%20range%20%26lt%3B20%20pg%5C%2FmL%29.%20PPP%20of%20ovarian%20origin%20was%20diagnosed%2C%20and%20tamoxifen%2020%20mg%20daily%20started.%20However%2C%20after%20only%20seven%20weeks%20height%20velocity%20escalated%20and%20breast%20development%20increased%20to%20B3-4%20with%20menorrhagia.%20Basal%5C%2F%20stimulated%20LH%20and%20FSH%20were%20still%20suppressed%20at%200.13%5C%2F0.25%20and%20%26lt%3B0.1%5C%2F%26lt%3B0.1%20mUI%5C%2FmL%20and%2C%20serum%20estradiol%20184%20pg%5C%2FmL.%20Repeat%20imaging%20now%20showed%20normal%20right%20ovary%20%28volume%201.8%20mL%29%20and%20a%20large%20left-sided%20vascular%20solid%5C%2Fcystic%20ovarian%20tumour%20which%20was%20excised%20%28weight%20850%20g%29.%20Histology%20showed%20JGCT%2C%20International%20Federation%20of%20Gynecology%20and%20Obstetrics%20stage%20IA.%20DNA%20from%20tumour%20tissue%20showed%20no%20mutation%20in%20GNAS%2C%20exon%203%20of%20AKT1%20%28which%20contains%20a%20mutational%20hotspot%29%20or%20FOXL2.%20The%20observation%20that%20bilateral%20ovarian%20activity%20progressed%20to%20unilateral%20development%20of%20JGCT%20in%20this%20patient%20is%20novel.%20This%20case%20highlights%20current%20uncertainties%20in%20the%20ontology%20of%20JGCT%2C%20and%20its%20possible%20relationship%20with%20MAS.%22%2C%22date%22%3A%222022-3-3%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.4274%5C%2Fjcrpe.galenos.2021.2021.0039%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fjag.journalagent.com%5C%2Fz4%5C%2Fdownload_fulltext.asp%3Fpdir%3Djcrpe%26plng%3Deng%26un%3DJCRPE-25733%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%2213085727%2C%2013085735%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-06-07T14%3A46%3A32Z%22%7D%7D%2C%7B%22key%22%3A%222CWIBU9M%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%20and%20Innan%22%2C%22parsedDate%22%3A%222022-03%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%2C%20%26amp%3B%20Innan%2C%20H.%20%282022%29.%20Pathogenic%20%26%23x201C%3Bgermline%26%23x201D%3B%20variants%20associated%20with%20myeloproliferative%20disorders%20in%20apparently%20normal%20individuals%3A%20Inherited%20or%20acquired%20genetic%20alterations%3F%20%26lt%3Bi%26gt%3BClinical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B101%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20371%26%23×2013%3B374.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.14104%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.14104%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Pathogenic%20%5C%22germline%5C%22%20variants%20associated%20with%20myeloproliferative%20disorders%20in%20apparently%20normal%20individuals%3A%20Inherited%20or%20acquired%20genetic%20alterations%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hideki%22%2C%22lastName%22%3A%22Innan%22%7D%5D%2C%22abstractNote%22%3A%22Myeloproliferative%20syndromes%20%28MPS%29%20are%20hematologic%20malignancies%20due%20to%20the%20expansion%20of%20an%20abnormal%20hematopoietic%20stem%20cell.%20They%20include%20chronic%20myeloid%20leukemia%20%28CML%29%20and%20non-CML%20MPS%20such%20as%20polycythemia%20vera%2C%20essential%20thrombocythemia%20and%20primary%20myelofibrosis.%20The%20latter%20are%20distinguished%20by%20somatic%20pathogenic%20variants%20affecting%20JAK2%2C%20CALR%2C%20or%20MPL%20genes.%20Apparent%20germline%20pathogenic%20variants%20have%20been%20reported%20in%20the%20general%20population.%20Here%2C%20we%20found%20that%20two%20gnomAD%20data-sets%20report%20more%20homozygotes%20than%20expected%20for%20the%20JAK2%20c.1849G%5Cu2009%26gt%3B%5Cu2009T%28Val617Phe%29%20variant.%20We%20propose%20that%20somatic%20gene%20conversion%20can%20explain%20the%20presence%20of%20those%20unexpected%20homozygotes%20in%20normal%20populations.%20Consistently%2C%20homozygous%20individuals%20are%20older%20than%2065%5Cu2009years.%20We%20also%20found%20a%20lower-than-expected%20frequency%20of%20the%20JAK2%20variant%20in%20younger%20individuals%20suggesting%20that%20somatic%20mutation%20can%20underlie%20its%20presence%20in%20%28at%20least%20some%29%20heterozygotes.%20Regarding%20pathogenic%20variants%20in%20MPL%20and%20CALR%2C%20they%20are%20also%20present%20in%20the%20gnomAD%20data-sets%20explored.%20However%2C%20we%20cannot%20conclude%20that%20such%20seemingly%20germline%20variants%20are%20in%20fact%20somatic%20alterations.%20These%20results%20suggest%20that%20apparently%20normal%20individuals%20bearing%20MPS-related%20variants%20can%20be%20subclinical%5C%2Fundiagnosed%20MPS%20cases%20of%20somatic%20origin.%20It%20would%20be%20interesting%20to%20assess%20the%20hematologic%20phenotype%20of%20such%20individuals%20and%20the%20presence%20of%20the%20relevant%20variants%20in%20other%20tissues.%22%2C%22date%22%3A%222022-03%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fcge.14104%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221399-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22PWBZYFEJ%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%20and%20Birchler%22%2C%22parsedDate%22%3A%222022-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%2C%20%26amp%3B%20Birchler%2C%20J.%20A.%20%282022%29.%20Gene-dosage%20issues%3A%20a%20recurrent%20theme%20in%20whole%20genome%20duplication%20events.%20%26lt%3Bi%26gt%3BTrends%20in%20Genetics%3A%20TIG%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B38%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%201%26%23×2013%3B3.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2021.06.006%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2021.06.006%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Gene-dosage%20issues%3A%20a%20recurrent%20theme%20in%20whole%20genome%20duplication%20events%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%20A.%22%2C%22lastName%22%3A%22Birchler%22%7D%5D%2C%22abstractNote%22%3A%22Two%20recent%20studies%20have%20addressed%20the%20long-term%20consequences%20of%20whole%20genome%20duplications%20%28WGD%29.%20Specifically%2C%20they%20analyzed%20transcriptomes%20of%20the%20plant%20Arabidopsis%20thaliana%20and%20of%20four%20salmonids%20to%20assess%20the%20impact%20of%20WGD%20on%20gene%20expression.%20These%20studies%20point%20to%20commonalities%20in%20gene%20expression%20adjustments%20after%20polyploidization%20that%20we%20outline%20and%20discuss%20below.%22%2C%22date%22%3A%222022-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tig.2021.06.006%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220168-9525%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%227NZX8SD4%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Pilsworth%20et%20al.%22%2C%22parsedDate%22%3A%222021-11%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPilsworth%2C%20J.%20A.%2C%20Todeschini%2C%20A.-L.%2C%20Neilson%2C%20S.%20J.%2C%20Cochrane%2C%20D.%20R.%2C%20Lai%2C%20D.%2C%20Anttonen%2C%20M.%2C%20Heikinheimo%2C%20M.%2C%20Huntsman%2C%20D.%20G.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282021%29.%20FOXL2%20in%20adult-type%20granulosa%20cell%20tumour%20of%20the%20ovary%3A%20oncogene%20or%20tumour%20suppressor%20gene%3F%20%26lt%3Bi%26gt%3BThe%20Journal%20of%20Pathology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B255%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20225%26%23×2013%3B231.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1002%5C%2Fpath.5771%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1002%5C%2Fpath.5771%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22FOXL2%20in%20adult-type%20granulosa%20cell%20tumour%20of%20the%20ovary%3A%20oncogene%20or%20tumour%20suppressor%20gene%3F%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%20A.%22%2C%22lastName%22%3A%22Pilsworth%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Samantha%20J.%22%2C%22lastName%22%3A%22Neilson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dawn%20R.%22%2C%22lastName%22%3A%22Cochrane%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%22%2C%22lastName%22%3A%22Lai%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mikko%22%2C%22lastName%22%3A%22Anttonen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Markku%22%2C%22lastName%22%3A%22Heikinheimo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22David%20G.%22%2C%22lastName%22%3A%22Huntsman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22A%20recurrent%20mutation%20in%20FOXL2%20%28c.402C%26gt%3BG%3B%20p.C134W%29%20is%20present%20in%20over%2095%25%20of%20adult-type%20granulosa%20cell%20tumours%20%28AGCTs%29.%20In%20contrast%2C%20various%20loss-of-function%20mutations%20in%20FOXL2%20lead%20to%20the%20development%20of%20blepharophimosis%2C%20ptosis%2C%20and%20epicanthus%20inversus%20syndrome%20%28BPES%29.%20BPES%20is%20characterised%20by%20an%20eyelid%20malformation%20often%20accompanied%20with%20primary%20ovarian%20insufficiency.%20Two%20recent%20studies%20suggest%20that%20FOXL2%20C402G%20is%20a%20gain-%20or%20change-of-function%20mutation%20with%20altered%20DNA-binding%20specificity.%20Another%20study%20proposes%20that%20FOXL2%20C402G%20is%20selectively%20targeted%20for%20degradation%2C%20inducing%20somatic%20haploinsufficiency%2C%20suggesting%20its%20role%20as%20a%20tumour%20suppressor.%20The%20latter%20study%20relies%20on%20data%20indicative%20of%20an%20FOXL2%20allelic%20imbalance%20in%20AGCTs.%20Here%20we%20present%20RNA-seq%20data%20as%20genetic%20evidence%20that%20no%20real%20allelic%20imbalance%20is%20observed%20at%20the%20transcriptomic%20level%20in%20AGCTs.%20Additionally%2C%20there%20is%20no%20loss%20of%20protein%20expression%20in%20tumours%20harbouring%20the%20mutated%20allele.%20These%20data%20and%20other%20features%20of%20this%20mutation%20compared%20to%20other%20oncogenes%20and%20tumour%20suppressor%20genes%20argue%20strongly%20against%20FOXL2%20being%20a%20tumour%20suppressor%20in%20this%20context.%20Given%20the%20likelihood%20that%20FOXL2%20C402G%20is%20oncogenic%2C%20targeting%20the%20variant%20protein%20or%20its%20downstream%20consequences%20is%20the%20most%20viable%20path%20forward%20to%20identifying%20an%20effective%20treatment%20for%20this%20cancer.%20%5Cu00a9%202021%20The%20Pathological%20Society%20of%20Great%20Britain%20and%20Ireland.%20Published%20by%20John%20Wiley%20%26amp%3B%20Sons%2C%20Ltd.%22%2C%22date%22%3A%222021-11%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1002%5C%2Fpath.5771%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221096-9896%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%223TJQ97IJ%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222021-09%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282021%29.%20A%20reply%20to%3A%20Longitudinal%20changes%20in%20the%20frequency%20of%20mosaic%20chromosome%20Y%20loss%20in%20peripheral%20blood%20cells%20of%20aging%20men%20varies%20profoundly%26%23xA0%3Bbetween%20individuals.%20%26lt%3Bi%26gt%3BEuropean%20Journal%20of%20Human%20Genetics%3A%20EJHG%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B29%26lt%3B%5C%2Fi%26gt%3B%289%29%2C%201321%26%23×2013%3B1322.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41431-020-00801-w%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41431-020-00801-w%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20reply%20to%3A%20Longitudinal%20changes%20in%20the%20frequency%20of%20mosaic%20chromosome%20Y%20loss%20in%20peripheral%20blood%20cells%20of%20aging%20men%20varies%20profoundly%5Cu00a0between%20individuals%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222021-09%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41431-020-00801-w%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221476-5438%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22593BGZDL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%20et%20al.%22%2C%22parsedDate%22%3A%222021-08-16%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%2C%20Pilsworth%2C%20J.%2C%20Todeschini%2C%20A.-L.%2C%20%26amp%3B%20Huntsman%2C%20D.%20%282021%29.%20Reply%20to%20%26%23x201C%3BAn%20alternative%20miRISC%20targets%20a%20cancer-associated%20coding%20sequence%20mutation%20in%20FOXL2.%26%23x201D%3B%20%26lt%3Bi%26gt%3BThe%20EMBO%20Journal%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B40%26lt%3B%5C%2Fi%26gt%3B%2816%29%2C%20e107517.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembj.2020107517%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembj.2020107517%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Reply%20to%20%5Cu201cAn%20alternative%20miRISC%20targets%20a%20cancer-associated%20coding%20sequence%20mutation%20in%20FOXL2%5Cu201d%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%22%2C%22lastName%22%3A%22Pilsworth%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22David%22%2C%22lastName%22%3A%22Huntsman%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222021-08-16%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.15252%5C%2Fembj.2020107517%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.15252%5C%2Fembj.2020107517%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220261-4189%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-06-07T14%3A50%3A05Z%22%7D%7D%2C%7B%22key%22%3A%225PTBIBDT%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Berm%5Cu00fadez-Guzm%5Cu00e1n%20and%20Veitia%22%2C%22parsedDate%22%3A%222021-07%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBerm%26%23xFA%3Bdez-Guzm%26%23xE1%3Bn%2C%20L.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282021%29.%20Insights%20into%20the%20pathogenicity%20of%20missense%20variants%20in%20the%20forkhead%20domain%20of%20FOX%20proteins%20underlying%20Mendelian%20disorders.%20%26lt%3Bi%26gt%3BHuman%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B140%26lt%3B%5C%2Fi%26gt%3B%287%29%2C%20999%26%23×2013%3B1010.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-021-02267-2%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-021-02267-2%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Insights%20into%20the%20pathogenicity%20of%20missense%20variants%20in%20the%20forkhead%20domain%20of%20FOX%20proteins%20underlying%20Mendelian%20disorders%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Luis%22%2C%22lastName%22%3A%22Berm%5Cu00fadez-Guzm%5Cu00e1n%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Forkhead%20box%20%28FOX%29%20proteins%20are%20members%20of%20a%20conserved%20family%20of%20transcription%20factors.%20Pathogenic%20variants%20in%20FOX%20genes%20have%20been%20shown%20to%20be%20responsible%20for%20several%20human%20genetic%20diseases.%20Here%2C%20we%20have%20studied%20the%20molecular%20and%20structural%20features%20of%20germline%20pathogenic%20variants%20in%20seven%20FOX%20proteins%20involved%20in%20Mendelian%20disorders%20and%20compared%20them%20with%20those%20of%20variants%20present%20in%20the%20general%20population%20%28gnomAD%29.%20Our%20study%20shows%20that%20the%20DNA-binding%20domain%20of%20FOX%20proteins%20is%20particularly%20sensitive%5Cu00a0to%20damaging%20variation%2C%20although%20some%20family%20members%20show%20greater%20mutational%20tolerance%20than%20others.%20Next%2C%20we%20set%20to%20demonstrate%20that%20this%20tolerance%20depends%20on%20the%20inheritance%20mode%20of%20FOX-linked%20disorders.%20Accordingly%2C%20genes%20whose%20variants%20underlie%20recessive%20conditions%20are%20supposed%20to%20have%20a%20greater%20tolerance%20to%20variation.%20This%20is%20what%20we%20found.%20As%20expected%2C%20variants%20responsible%20for%20disorders%20with%20a%20dominant%20inheritance%20pattern%20show%20a%20higher%20degree%20of%20pathogenicity%20compared%20to%20those%20segregating%20in%20the%20general%20population.%20Moreover%2C%20we%20show%20that%20pathogenic%20and%20likely%20pathogenic%20variants%20tend%20to%20affect%20mutually%20exclusive%20sites%20with%20respect%20to%20those%20reported%20in%20gnomAD.%20The%20former%20also%20tend%20to%20affect%20sites%20with%20lower%20solvent%20exposure%20and%20a%20higher%20degree%20of%20conservation.%20Our%20results%20show%20the%20value%20of%20using%20publicly%20available%20databases%20and%20bioinformatics%20to%20gain%20insights%20into%20the%20molecular%20and%20structural%20bases%20of%20disease-causing%20genetic%20variation.%22%2C%22date%22%3A%222021-07%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00439-021-02267-2%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221432-1203%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22BJESICF2%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Herman%20et%20al.%22%2C%22parsedDate%22%3A%222021-05%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BHerman%2C%20L.%2C%20Todeschini%2C%20A.-L.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282021%29.%20Forkhead%20Transcription%20Factors%20in%20Health%20and%20Disease.%20%26lt%3Bi%26gt%3BTrends%20in%20Genetics%3A%20TIG%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B37%26lt%3B%5C%2Fi%26gt%3B%285%29%2C%20460%26%23×2013%3B475.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2020.11.003%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2020.11.003%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Forkhead%20Transcription%20Factors%20in%20Health%20and%20Disease%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laetitia%22%2C%22lastName%22%3A%22Herman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Forkhead%20box%20%28FOX%29%20proteins%20belong%20to%20an%20evolutionarily%20conserved%20family%20of%20transcription%20factors%20that%20has%20evolved%20by%20gene%5C%2Fgenome%20duplication.%20FOX%20family%20members%20have%20undergone%20sequence%20and%20regulatory%20diversification.%20However%2C%20they%20have%20retained%20some%20degree%20of%20functional%20redundancy%2C%20in%20addition%20to%20playing%20specific%20roles%2C%20both%20during%20development%20and%20in%20the%20adult.%20Genetic%20alterations%20or%20misregulation%20of%20FOX%20genes%20underlie%20human%20genetic%20diseases%2C%20cancer%2C%20and%5C%2For%20aging.%20In%20this%20review%2C%20we%20provide%20an%20updated%20overview%20of%20the%20main%20characteristics%20of%20the%20members%20of%20this%20family%2C%20in%20terms%20of%20breadth%20of%20expression%2C%20protein%20domain%20composition%2C%20evolution%2C%20and%20function.%22%2C%22date%22%3A%222021-05%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tig.2020.11.003%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220168-9525%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22RYZRMAGG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Innan%20et%20al.%22%2C%22parsedDate%22%3A%222021-05%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BInnan%2C%20H.%2C%20Vaiman%2C%20D.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282021%29.%20Predictable%20increase%20in%20female%20reproductive%20window%3A%20A%20simple%20model%20connecting%20age%20of%20reproduction%2C%20menopause%2C%20and%20longevity.%20%26lt%3Bi%26gt%3BBioEssays%3A%20News%20and%20Reviews%20in%20Molecular%2C%20Cellular%20and%20Developmental%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B43%26lt%3B%5C%2Fi%26gt%3B%285%29%2C%20e2000233.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1002%5C%2Fbies.202000233%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1002%5C%2Fbies.202000233%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Predictable%20increase%20in%20female%20reproductive%20window%3A%20A%20simple%20model%20connecting%20age%20of%20reproduction%2C%20menopause%2C%20and%20longevity%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hideki%22%2C%22lastName%22%3A%22Innan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%22%2C%22lastName%22%3A%22Vaiman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22With%20the%20ever-increasing%20lifespan%20along%20with%20societal%20changes%2C%20women%20can%20marry%20and%20procreate%20later%20than%20in%20previous%20centuries.%20However%2C%20pathogenic%20genetic%20variants%20segregating%20in%20the%20population%20can%20lead%20to%20female%20subfertility%20or%20infertility%20well%20before%20the%20average%20age%20of%20normal%20menopause%2C%20leading%20to%20counter-selection%20of%20such%20deleterious%20alleles.%20In%20reviewing%20this%20field%2C%20we%20speculate%20that%20a%20logical%20consequence%20would%20be%20the%20later%20occurrence%20of%20menopause%20and%20the%20extension%20of%20women%26%23039%3Bs%20reproductive%20lifespan.%20We%20illustrate%20this%20point%20with%20a%20simple%20model%20that%20applies%20to%20other%20variants%20that%20contribute%20to%20female%20infertility%2C%20including%20epigenetic%20variation.%20We%20also%20consider%20the%20effect%20of%20medical%20interventions%20and%20lifestyle.%22%2C%22date%22%3A%222021-05%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1002%5C%2Fbies.202000233%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221521-1878%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22XNRNB3PS%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Herman%20et%20al.%22%2C%22parsedDate%22%3A%222021-04%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BHerman%2C%20L.%2C%20Legois%2C%20B.%2C%20Todeschini%2C%20A.-L.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282021%29.%20Genomic%20exploration%20of%20the%20targets%20of%20FOXL2%20and%20ESR2%20unveils%20their%20implication%20in%20cell%20migration%2C%20invasion%2C%20and%20adhesion.%20%26lt%3Bi%26gt%3BFASEB%20Journal%3A%20Official%20Publication%20of%20the%20Federation%20of%20American%20Societies%20for%20Experimental%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B35%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20e21355.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1096%5C%2Ffj.202002444R%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1096%5C%2Ffj.202002444R%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Genomic%20exploration%20of%20the%20targets%20of%20FOXL2%20and%20ESR2%20unveils%20their%20implication%20in%20cell%20migration%2C%20invasion%2C%20and%20adhesion%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laetitia%22%2C%22lastName%22%3A%22Herman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B%5Cu00e9rang%5Cu00e8re%22%2C%22lastName%22%3A%22Legois%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22FOXL2%20and%20ESR2%20are%20key%20transcriptional%20regulators%20in%20ovarian%20granulosa%20cells.%20To%20explore%20their%20transcriptional%20roles%20and%20their%20interplay%2C%20we%20have%20depleted%20Foxl2%20and%20Esr2%20in%20mouse%20primary%20granulosa%20cells%20to%20assess%20their%20ability%20to%20bind%20their%20targets%20and%5C%2For%20to%20modulate%20gene%20expression%20and%20cellular%20functions.%20We%20show%20that%20FOXL2%20is%20involved%20in%20a%20large%20number%20of%20regulatory%20actions%20essential%20for%20the%20maintenance%20of%20granulosa%20cell%20fate.%20A%20parallel%20ChIP-seq%20analysis%20showed%20that%20FOXL2%20mainly%20binds%20to%20sites%20located%20in%20intergenic%20regions%20quite%20far%20from%20its%20targets.%20A%20bioinformatic%20analysis%20demonstrated%20that%20FOXL2-activated%20genes%20were%20enriched%20in%20peaks%20associated%20with%20the%20H3K27ac%20mark%2C%20whereas%20FOXL2-repressed%20genes%20were%20not%2C%20suggesting%20that%20FOXL2%20can%20activate%20transcription%20through%20binding%20to%20enhancer%20sites.%20We%20also%20identified%20about%20500%20deregulated%20genes%20upon%20Esr2%20silencing%2C%20of%20which%20one%20third%20are%20also%20targets%20of%20FOXL2.%20We%20provide%20evidence%20showing%20that%20both%20factors%20modulate%2C%20through%20a%20coherent%20feed-forward%20loop%2C%20a%20number%20of%20common%20targets.%20Many%20of%20the%20FOXL2%5C%2FESR2%20targets%20are%20involved%20in%20cell%20motility%20and%2C%20consistently%2C%20granulosa%20cells%20depleted%20for%20either%20Foxl2%20or%20Esr2%20exhibit%20decreased%20migration%2C%20invasion%20and%20adhesion.%20This%20effect%20is%20paralleled%20by%20the%20depletion%20of%20their%20target%20Phactr1%2C%20involved%20in%20actin%20cytoskeleton%20dynamics.%20Our%20analysis%20expands%20the%20number%20of%20direct%20and%20indirect%20transcriptional%20targets%20of%20both%20FOXL2%20and%20ESR2%2C%20which%20deserve%20investigation%20in%20the%20context%20of%20adult-type%20granulosa%20cell%20tumors%20whose%20molecular%20diagnostic%20hallmark%20is%20the%20presence%20of%20the%20C134W%20FOXL2%20pathogenic%20variant.%22%2C%22date%22%3A%222021-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1096%5C%2Ffj.202002444R%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221530-6860%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22C64V8THN%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Caburet%20et%20al.%22%2C%22parsedDate%22%3A%222021-02-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BCaburet%2C%20S.%2C%20Heddar%2C%20A.%2C%20Dardillac%2C%20E.%2C%20Creux%2C%20H.%2C%20Lambert%2C%20M.%2C%20Messiaen%2C%20S.%2C%20Tourpin%2C%20S.%2C%20Livera%2C%20G.%2C%20Lopez%2C%20B.%20S.%2C%20%26amp%3B%20Misrahi%2C%20M.%20%282021%29.%20Homozygous%20hypomorphic%20BRCA2%20variant%20in%20primary%20ovarian%20insufficiency%20without%20cancer%20or%20Fanconi%20anaemia%20trait.%20%26lt%3Bi%26gt%3BJournal%20of%20Medical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B58%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20125%26%23×2013%3B134.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1136%5C%2Fjmedgenet-2019-106672%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1136%5C%2Fjmedgenet-2019-106672%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Homozygous%20hypomorphic%20BRCA2%20variant%20in%20primary%20ovarian%20insufficiency%20without%20cancer%20or%20Fanconi%20anaemia%20trait%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Caburet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Abdelkader%22%2C%22lastName%22%3A%22Heddar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elodie%22%2C%22lastName%22%3A%22Dardillac%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22H%5Cu00e9l%5Cu00e9ne%22%2C%22lastName%22%3A%22Creux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marie%22%2C%22lastName%22%3A%22Lambert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e9bastien%22%2C%22lastName%22%3A%22Messiaen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sophie%22%2C%22lastName%22%3A%22Tourpin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gabriel%22%2C%22lastName%22%3A%22Livera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bernard%20S.%22%2C%22lastName%22%3A%22Lopez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Micheline%22%2C%22lastName%22%3A%22Misrahi%22%7D%5D%2C%22abstractNote%22%3A%22Background%20Primary%20ovarian%20insufficiency%20%28POI%29%20affects%201%25%20of%20women%20under%2040%20years%20and%20is%20a%20public%20health%20problem.%20The%20genetic%20causes%20of%20POI%20are%20highly%20heterogeneous%20with%20isolated%20or%20syndromic%20forms.%20Recently%2C%20variants%20in%20genes%20involved%20in%20DNA%20repair%20have%20been%20shown%20to%20cause%20POI.%20Notably%2C%20syndromic%20POI%20with%20Fanconi%20anaemia%20%28FA%29%20traits%20related%20to%20biallelic%20BRCA2%20truncated%20variants%20has%20been%20reported.%20Here%2C%20we%20report%20a%20novel%20phenotype%20of%20isolated%20POI%20with%20a%20BRCA2%20variant%20in%20a%20consanguineous%20Turkish%20family.%5CnMethods%20Exome%20sequencing%20%28ES%29%20was%20performed%20in%20the%20patient.%20We%20also%20performed%20functional%20studies%2C%20including%20a%20homologous%20recombination%20%28HR%29%20test%2C%20cell%20proliferation%2C%20radiation-induced%20RAD51%20foci%20formation%20assays%20and%20chromosome%20breakage%20studies%20in%20primary%20and%20lymphoblastoid%20immortalised%20cells.%20The%20expression%20of%20BRCA2%20in%20human%20foetal%20ovaries%20was%20studied.%5CnResults%20ES%20identified%20a%20homozygous%20missense%20c.8524C%26gt%3BT%5C%2Fp.R2842C-BRCA2%20variant.%20BRCA2%20defects%20induce%20cancer%20predisposition%20and%20FA.%20Remarkably%2C%20neither%20the%20patient%20nor%20her%20family%20exhibited%20somatic%20pathologies.%20The%20patient%5Cu2019s%20cells%20showed%20intermediate%20levels%20of%20chromosomal%20breaks%2C%20cell%20proliferation%20and%20radiation-induced%20RAD51%20foci%20formation%20compared%20with%20controls%20and%20FA%20cells.%20R2842C-BRCA2%20only%20partially%20complemented%20HR%20efficiency%20compared%20with%20wild%20type-BRCA2.%20BRCA2%20is%20expressed%20in%20human%20foetal%20ovaries%20in%20pachytene%20stage%20oocytes%2C%20when%20meiotic%20HR%20occurs.%5CnConclusion%20We%20describe%20the%20functional%20assessment%20of%20a%20homozygous%20hypomorphic%20BRCA2%20variant%20in%20a%20patient%20with%20POI%20without%20cancer%20or%20FA%20trait.%20Our%20findings%20extend%20the%20phenotype%20of%20BRCA2%20biallelic%20alterations%20to%20fully%20isolated%20POI.%20This%20study%20has%20a%20major%20impact%20on%20the%20management%20and%20genetic%20counselling%20of%20patients%20with%20POI.%22%2C%22date%22%3A%222021%5C%2F02%5C%2F01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1136%5C%2Fjmedgenet-2019-106672%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fjmg.bmj.com%5C%2Fcontent%5C%2F58%5C%2F2%5C%2F125%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220022-2593%2C%201468-6244%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-06-07T14%3A52%3A29Z%22%7D%7D%2C%7B%22key%22%3A%22LQK2R9PX%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222021-02%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282021%29.%20Clinical%20Genetics%20paving%20the%20way%20to%20the%20future.%20%26lt%3Bi%26gt%3BClinical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B99%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20217%26%23×2013%3B218.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13899%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13899%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Clinical%20Genetics%20paving%20the%20way%20to%20the%20future%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222021-02%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fcge.13899%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221399-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22TSDPAYDN%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Birchler%20and%20Veitia%22%2C%22parsedDate%22%3A%222021%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBirchler%2C%20J.%20A.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282021%29.%20One%20Hundred%20Years%20of%20Gene%20Balance%3A%20How%20Stoichiometric%20Issues%20Affect%20Gene%20Expression%2C%20Genome%20Evolution%2C%20and%20Quantitative%20Traits.%20%26lt%3Bi%26gt%3BCytogenetic%20and%20Genome%20Research%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B161%26lt%3B%5C%2Fi%26gt%3B%2810%26%23×2013%3B11%29%2C%20529%26%23×2013%3B550.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1159%5C%2F000519592%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1159%5C%2F000519592%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22One%20Hundred%20Years%20of%20Gene%20Balance%3A%20How%20Stoichiometric%20Issues%20Affect%20Gene%20Expression%2C%20Genome%20Evolution%2C%20and%20Quantitative%20Traits%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%20A.%22%2C%22lastName%22%3A%22Birchler%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22A%20century%20ago%20experiments%20with%20the%20flowering%20plant%20Datura%20stramonium%20and%20the%20fruit%20fly%20Drosophila%20melanogaster%20revealed%20that%20adding%20an%20extra%20chromosome%20to%20a%20karyotype%20was%20much%20more%20detrimental%20than%20adding%20a%20whole%20set%20of%20chromosomes.%20This%20phenomenon%20was%20referred%20to%20as%20gene%20balance%20and%20has%20been%20recapitulated%20across%20eukaryotic%20species.%20Here%2C%20we%20retrace%20some%20developments%20in%20this%20field.%20Molecular%20studies%20suggest%20that%20the%20basis%20of%20balance%20involves%20stoichiometric%20relationships%20of%20multi-component%20interactions.%20This%20concept%20has%20implication%20for%20the%20mechanisms%20controlling%20gene%20expression%2C%20genome%20evolution%2C%20sex%20chromosome%20evolution%5C%2Fdosage%20compensation%2C%20speciation%20mechanisms%2C%20and%20the%20underlying%20genetics%20of%20quantitative%20traits.%22%2C%22date%22%3A%222021%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1159%5C%2F000519592%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221424-859X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22PW39C93M%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Picolo%20et%20al.%22%2C%22parsedDate%22%3A%222021%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPicolo%2C%20F.%2C%20Grandchamp%2C%20A.%2C%20Pi%26%23xE9%3Bgu%2C%20B.%2C%20Rolland%2C%20A.%20D.%2C%20Veitia%2C%20R.%20A.%2C%20%26amp%3B%20Monget%2C%20P.%20%282021%29.%20Genes%20Encoding%20Teleost%20Orthologs%20of%20Human%20Haploinsufficient%20and%20Monoallelically%20Expressed%20Genes%20Remain%20in%20Duplicate%20More%20Frequently%20Than%20the%20Whole%20Genome.%20%26lt%3Bi%26gt%3BInternational%20Journal%20of%20Genomics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B2021%26lt%3B%5C%2Fi%26gt%3B%2C%209028667.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1155%5C%2F2021%5C%2F9028667%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1155%5C%2F2021%5C%2F9028667%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Genes%20Encoding%20Teleost%20Orthologs%20of%20Human%20Haploinsufficient%20and%20Monoallelically%20Expressed%20Genes%20Remain%20in%20Duplicate%20More%20Frequently%20Than%20the%20Whole%20Genome%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Floriane%22%2C%22lastName%22%3A%22Picolo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Grandchamp%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Beno%5Cu00eet%22%2C%22lastName%22%3A%22Pi%5Cu00e9gu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Antoine%20D.%22%2C%22lastName%22%3A%22Rolland%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Philippe%22%2C%22lastName%22%3A%22Monget%22%7D%5D%2C%22abstractNote%22%3A%22Gene%20dosage%20is%20an%20important%20issue%20both%20in%20cell%20and%20evolutionary%20biology.%20Most%20genes%20are%20present%20in%20two%20copies%20or%20alleles%20in%20diploid%20eukariotic%20cells.%20The%20most%20outstanding%20exception%20is%20monoallelic%20gene%20expression%20%28MA%29%20that%20concerns%20genes%20localized%20on%20the%20X%20chromosome%20or%20in%20regions%20undergoing%20parental%20imprinting%20in%20eutherians%2C%20and%20many%20other%20genes%20scattered%20throughout%20the%20genome.%20In%20diploids%2C%20haploinsufficiency%20%28HI%29%20implies%20that%20a%20single%20functional%20copy%20of%20a%20gene%20in%20a%20diploid%20organism%20is%20insufficient%20to%20ensure%20a%20normal%20biological%20function.%20One%20of%20the%20most%20important%20mechanisms%20ensuring%20functional%20innovation%20during%20evolution%20is%20whole%20genome%20duplication%20%28WGD%29.%20In%20addition%20to%20the%20two%20WGDs%20that%20have%20occurred%20in%20vertebrate%20genomes%2C%20the%20teleost%20genomes%20underwent%20an%20additional%20WGD%2C%20after%20their%20divergence%20from%20tetrapods.%20In%20the%20present%20work%2C%20we%20have%20studied%20on%2057%20teleost%20species%20whether%20the%20orthologs%20of%20human%20MA%20or%20HI%20genes%20remain%20more%20frequently%20in%20duplicates%20or%20returned%20more%20frequently%20in%20singleton%20than%20the%20rest%20of%20the%20genome.%20Our%20results%20show%20that%20the%20teleost%20orthologs%20of%20HI%20human%20genes%20remained%20more%20frequently%20in%20duplicate%20than%20the%20rest%20of%20the%20genome%20in%20all%20of%20the%20teleost%20species%20studied.%20No%20signal%20was%20observed%20for%20the%20orthologs%20of%20genes%20mapping%20to%20the%20human%20X%20chromosome%20or%20subjected%20to%20parental%20imprinting.%20Surprisingly%2C%20the%20teleost%20orthologs%20of%20the%20other%20human%20MA%20genes%20remained%20in%20duplicate%20more%20frequently%20than%20the%20rest%20of%20the%20genome%20for%20most%20teleost%20species.%20These%20results%20suggest%20that%20the%20teleost%20orthologs%20of%20MA%20and%20HI%20human%20genes%20also%20undergo%20selective%20pressures%20either%20related%20to%20absolute%20protein%20amounts%20and%5C%2For%20of%20dosage%20balance%20issues.%20However%2C%20these%20constraints%20seem%20to%20be%20different%20for%20MA%20genes%20in%20teleost%20in%20comparison%20with%20human%20genomes.%22%2C%22date%22%3A%222021%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1155%5C%2F2021%5C%2F9028667%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222314-4378%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22SUJJ9LCW%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Elzaiat%20et%20al.%22%2C%22parsedDate%22%3A%222020-11%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BElzaiat%2C%20M.%2C%20Flatters%2C%20D.%2C%20Sierra-D%26%23xED%3Baz%2C%20D.%20C.%2C%20Legois%2C%20B.%2C%20Laissue%2C%20P.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282020%29.%20DHH%20pathogenic%20variants%20involved%20in%2046%2CXY%20disorders%20of%20sex%20development%20differentially%20impact%20protein%20self-cleavage%20and%20structural%20conformation.%20%26lt%3Bi%26gt%3BHuman%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B139%26lt%3B%5C%2Fi%26gt%3B%2811%29%2C%201455%26%23×2013%3B1470.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-020-02189-5%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-020-02189-5%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22DHH%20pathogenic%20variants%20involved%20in%2046%2CXY%20disorders%20of%20sex%20development%20differentially%20impact%20protein%20self-cleavage%20and%20structural%20conformation%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ma%5Cu00ebva%22%2C%22lastName%22%3A%22Elzaiat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Delphine%22%2C%22lastName%22%3A%22Flatters%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Diana%20Carolina%22%2C%22lastName%22%3A%22Sierra-D%5Cu00edaz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Berang%5Cu00e8re%22%2C%22lastName%22%3A%22Legois%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paul%22%2C%22lastName%22%3A%22Laissue%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22In%20humans%2C%20pathogenic%20variants%20in%20the%20DHH%20gene%20underlie%20cases%20of%2046%2CXY%20gonadal%20dysgenesis.%20DHH%20is%20part%20of%20the%20Hedgehog%20family%20of%20proteins%2C%20which%20require%20extensive%20processing%2C%20including%20self-cleavage%20of%20the%20precursor%20for%20efficient%20signalling.%20In%20our%20work%2C%20we%20have%20assessed%20the%20effect%20of%20several%20human%20DHH%20pathogenic%20variants%20involved%20in%20recessive%20complete%20or%20partial%20gonadal%20dysgenesis%2C%20on%20protein%20processing%20and%20sub-cellular%20localization.%20We%20found%20that%20a%20subset%20of%20variants%20was%20unable%20to%20perform%20self-cleavage%2C%20which%20correlated%20albeit%20not%20perfectly%20with%20an%20altered%20subcellular%20localization%20of%20the%20resulting%20proteins.%20For%20the%20processing-proficient%20variants%2C%20we%20used%20structural%20modelling%20tools%20and%20molecular%20dynamic%20%28MD%29%20simulations%20to%20predict%20the%20potential%20impact%20of%20the%20variants%20on%20protein%20conformation%20and%5C%2For%20interaction%20with%20partners.%20Our%20study%20contributes%20to%20a%20better%20understanding%20of%20the%20molecular%20mechanisms%20involved%20in%20DHH%20dysfunction%20leading%20to%2046%2CXY%20disorders%20of%20sex%20development.%22%2C%22date%22%3A%222020-11%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00439-020-02189-5%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221432-1203%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22EGEENALP%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Alvarez-Mora%20et%20al.%22%2C%22parsedDate%22%3A%222020-09%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BAlvarez-Mora%2C%20M.%20I.%2C%20Todeschini%2C%20A.-L.%2C%20Caburet%2C%20S.%2C%20Perets%2C%20L.%20P.%2C%20Mila%2C%20M.%2C%20Younis%2C%20J.%20S.%2C%20Shalev%2C%20S.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282020%29.%20An%20exome-wide%20exploration%20of%20cases%20of%20primary%20ovarian%20insufficiency%20uncovers%20novel%20sequence%20variants%20and%20candidate%20genes.%20%26lt%3Bi%26gt%3BClinical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B98%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20293%26%23×2013%3B298.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13803%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13803%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22An%20exome-wide%20exploration%20of%20cases%20of%20primary%20ovarian%20insufficiency%20uncovers%20novel%20sequence%20variants%20and%20candidate%20genes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maria%20Isabel%22%2C%22lastName%22%3A%22Alvarez-Mora%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Caburet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lilach%20Peled%22%2C%22lastName%22%3A%22Perets%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Montserrat%22%2C%22lastName%22%3A%22Mila%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Johnny%20S.%22%2C%22lastName%22%3A%22Younis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stavit%22%2C%22lastName%22%3A%22Shalev%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Primary%20ovarian%20insufficiency%20%28POI%29%20implies%20the%20cessation%20of%20menstruation%20for%20several%20months%20in%20women%20before%20the%20age%20of%2040%5Cu2009years%20and%20is%20a%20major%20cause%20of%20infertility.%20The%20study%20of%20the%20contribution%20of%20genetic%20factors%20to%20POI%20has%20been%20fueled%20by%20the%20use%20of%20whole%20exome%20sequencing%20%28WES%29.%20Here%2C%20to%20uncover%20novel%20causative%20pathogenic%20variants%20and%20risk%20alleles%2C%20WES%20has%20been%20performed%20in%2012%20patients%20with%20familial%20POI%20%28eight%20unrelated%20index%20cases%20and%20two%20pairs%20of%20sisters%29%20and%20six%20women%20with%20early%20menopause%20and%20family%20history%20of%20POI%20%28four%20index%20cases%20and%20one%20pair%20of%20sisters%29.%20Likely%20causative%20variants%20in%20NR5A1%20and%20MCM9%20genes%20were%20identified%20as%20well%20as%20a%20variant%20in%20INHA%20that%20requires%20further%20investigation.%20Moreover%2C%20we%20have%20identified%20more%20than%20one%20candidate%20variant%20in%203%20out%20of%2015%20familial%20cases.%20Taken%20together%2C%20our%20results%20highlight%20the%20genetic%20heterogeneity%20of%20POI%20and%20early%20menopause%20and%20support%20the%20hypothesis%20of%20an%20oligogenic%20inheritance%20of%20such%20conditions%2C%20in%20addition%20to%20monogenic%20inheritance.%22%2C%22date%22%3A%222020-09%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fcge.13803%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221399-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%229Q8ZZECZ%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Felipe-Medina%20et%20al.%22%2C%22parsedDate%22%3A%222020-08-26%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BFelipe-Medina%2C%20N.%2C%20Caburet%2C%20S.%2C%20S%26%23xE1%3Bnchez-S%26%23xE1%3Bez%2C%20F.%2C%20Condezo%2C%20Y.%20B.%2C%20de%20Rooij%2C%20D.%20G.%2C%20G%26%23xF3%3Bmez-H%2C%20L.%2C%20Garcia-Valiente%2C%20R.%2C%20Todeschini%2C%20A.%20L.%2C%20Duque%2C%20P.%2C%20S%26%23xE1%3Bnchez-Martin%2C%20M.%20A.%2C%20Shalev%2C%20S.%20A.%2C%20Llano%2C%20E.%2C%20Veitia%2C%20R.%20A.%2C%20%26amp%3B%20Pend%26%23xE1%3Bs%2C%20A.%20M.%20%282020%29.%20A%20missense%20in%20HSF2BP%20causing%20primary%20ovarian%20insufficiency%20affects%20meiotic%20recombination%20by%20its%20novel%20interactor%20C19ORF57%5C%2FBRME1.%20%26lt%3Bi%26gt%3BeLife%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2C%20e56996.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.56996%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.56996%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20missense%20in%20HSF2BP%20causing%20primary%20ovarian%20insufficiency%20affects%20meiotic%20recombination%20by%20its%20novel%20interactor%20C19ORF57%5C%2FBRME1%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Natalia%22%2C%22lastName%22%3A%22Felipe-Medina%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Caburet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fernando%22%2C%22lastName%22%3A%22S%5Cu00e1nchez-S%5Cu00e1ez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yazmine%20B.%22%2C%22lastName%22%3A%22Condezo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dirk%20G.%22%2C%22lastName%22%3A%22de%20Rooij%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22G%5Cu00f3mez-H%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rodrigo%22%2C%22lastName%22%3A%22Garcia-Valiente%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne%20Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paloma%22%2C%22lastName%22%3A%22Duque%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Manuel%20Adolfo%22%2C%22lastName%22%3A%22S%5Cu00e1nchez-Martin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stavit%20A.%22%2C%22lastName%22%3A%22Shalev%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elena%22%2C%22lastName%22%3A%22Llano%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alberto%20M.%22%2C%22lastName%22%3A%22Pend%5Cu00e1s%22%7D%5D%2C%22abstractNote%22%3A%22Primary%20Ovarian%20Insufficiency%20%28POI%29%20is%20a%20major%20cause%20of%20infertility%2C%20but%20its%20etiology%20remains%20poorly%20understood.%20Using%20whole-exome%20sequencing%20in%20a%20family%20with%20three%20cases%20of%20POI%2C%20we%20identified%20the%20candidate%20missense%20variant%20S167L%20in%20HSF2BP%2C%20an%20essential%20meiotic%20gene.%20Functional%20analysis%20of%20the%20HSF2BP-S167L%20variant%20in%20mouse%20showed%20that%20it%20behaves%20as%20a%20hypomorphic%20allele%20compared%20to%20a%20new%20loss-of-function%20%28knock-out%29%20mouse%20model.%20Hsf2bpS167L%5C%2FS167L%20females%20show%20reduced%20fertility%20with%20smaller%20litter%20sizes.%20To%20obtain%20mechanistic%20insights%2C%20we%20identified%20C19ORF57%5C%2FBRME1%20as%20a%20strong%20interactor%20and%20stabilizer%20of%20HSF2BP%20and%20showed%20that%20the%20BRME1%5C%2FHSF2BP%20protein%20complex%20co-immunoprecipitates%20with%20BRCA2%2C%20RAD51%2C%20RPA%20and%20PALB2.%20Meiocytes%20bearing%20the%20HSF2BP-S167L%20variant%20showed%20a%20strongly%20decreased%20staining%20of%20both%20HSF2BP%20and%20BRME1%20at%20the%20recombination%20nodules%20and%20a%20reduced%20number%20of%20the%20foci%20formed%20by%20the%20recombinases%20RAD51%5C%2FDMC1%2C%20thus%20leading%20to%20a%20lower%20frequency%20of%20crossovers.%20Our%20results%20provide%20insights%20into%20the%20molecular%20mechanism%20of%20HSF2BP-S167L%20in%20human%20ovarian%20insufficiency%20and%20sub%28in%29fertility.%22%2C%22date%22%3A%222020-08-26%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.56996%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222050-084X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22L2WRJBDV%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Govindaraju%20et%20al.%22%2C%22parsedDate%22%3A%222020-06%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BGovindaraju%2C%20D.%20R.%2C%20Innan%2C%20H.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282020%29.%20The%20Muller%26%23×2019%3Bs%20Ratchet%20and%20Aging.%20%26lt%3Bi%26gt%3BTrends%20in%20Genetics%3A%20TIG%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B36%26lt%3B%5C%2Fi%26gt%3B%286%29%2C%20395%26%23×2013%3B402.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2020.02.004%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2020.02.004%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20Muller%27s%20Ratchet%20and%20Aging%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Diddahally%20R.%22%2C%22lastName%22%3A%22Govindaraju%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hideki%22%2C%22lastName%22%3A%22Innan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Aging%20entails%20an%20irreversible%20deceleration%20of%20physiological%20processes%2C%20altered%20metabolic%20activities%2C%20and%20a%20decline%20of%20the%20integrity%20of%20tissues%2C%20organs%2C%20and%20organ%20systems.%20The%20accumulation%20of%20alterations%20in%20the%20genetic%20and%20epigenetic%20spaces%20has%20been%20proposed%20as%20an%20explanation%20for%20aging.%20They%20result%2C%20at%20least%20in%20part%2C%20from%20DNA%20replication%20and%20chromosome%20segregation%20errors%20due%20to%20cell%20division%20during%20development%2C%20growth%2C%20renewal%2C%20and%20repair.%20Such%20deleterious%20alterations%2C%20including%20epigenetic%20drift%2C%20irreversibly%20accumulate%20in%20a%20stepwise%2C%20ratchet-like%20manner%20and%20reduce%20cellular%20fitness%2C%20similar%20to%20the%20process%20known%20as%20Muller%26%23039%3Bs%20ratchet.%20Here%2C%20we%20revisit%20the%20Muller%26%23039%3Bs%20ratchet%20principle%20applied%20to%20the%20aging%20of%20somatic%20cell%20populations%20and%20discuss%20the%20implications%20for%20understanding%20the%20origins%20of%20senescence%2C%20frailty%2C%20and%20morbidity.%22%2C%22date%22%3A%222020-06%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tig.2020.02.004%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220168-9525%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22V4JEPXYQ%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222020-04%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282020%29.%20Primary%20ovarian%20insufficiency%2C%20meiosis%20and%20DNA%20repair.%20%26lt%3Bi%26gt%3BBiomedical%20Journal%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B43%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20115%26%23×2013%3B123.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.bj.2020.03.005%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.bj.2020.03.005%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Primary%20ovarian%20insufficiency%2C%20meiosis%20and%20DNA%20repair%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Premature%20ovarian%20insufficiency%20%28POI%29%20is%20a%20major%20cause%20of%20female%20infertility.%20It%20is%20a%20heterogeneous%20disease%20that%20affects%20about%201%25%20of%20women%20under%2040%20years%20of%20age.%20POI%20may%20be%20due%20to%20abnormal%20follicle%20stock%20formation%2C%20increased%20follicular%20atresia%2C%20impaired%20recruitment%20of%20dominant%20follicles%2C%20blocked%20follicular%20maturation%20or%20rapid%20depletion%20of%20the%20follicular%20stock.%20It%20remains%20idiopathic%20in%20most%20cases%20but%20the%20existence%20of%20familial%20cases%20shows%20that%20it%20can%20have%20a%20genetic%20origin.%20Next%20generation%20sequencing%20%28NGS%29%20strategies%20have%20allowed%20the%20identification%20of%20new%20genes%20involved%20in%20the%20etiology%20of%20POI.%20Here%2C%20I%20briefly%20describe%20some%20studies%20demonstrating%20that%20pathogenic%20variants%20in%20%26%23039%3BDNA%20repair%20and%20meiotic%20genes%26%23039%3B%20underlie%20POI.%20Some%20of%20the%20examples%20show%20the%20power%20of%20the%20combination%20of%20classical%20genetics%20and%20NGS%20in%20the%20discovery%20of%20novel%20%26%23039%3BPOI%20genes%26%23039%3B.%22%2C%22date%22%3A%222020-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.bj.2020.03.005%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222320-2890%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-06-07T14%3A52%3A59Z%22%7D%7D%2C%7B%22key%22%3A%22MGRGFYMR%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Innan%20et%20al.%22%2C%22parsedDate%22%3A%222020-03-01%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BInnan%2C%20H.%2C%20Veitia%2C%20R.%2C%20%26amp%3B%20Govindaraju%2C%20D.%20R.%20%282020%29.%20Genetic%20and%20epigenetic%20Muller%26%23×2019%3Bs%20ratchet%20as%20a%20mechanism%20of%20frailty%20and%20morbidity%20during%20aging%3A%20a%20demographic%20genetic%20model.%20%26lt%3Bi%26gt%3BHuman%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B139%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20409%26%23×2013%3B420.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-019-02067-9%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-019-02067-9%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Genetic%20and%20epigenetic%20Muller%5Cu2019s%20ratchet%20as%20a%20mechanism%20of%20frailty%20and%20morbidity%20during%20aging%3A%20a%20demographic%20genetic%20model%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hideki%22%2C%22lastName%22%3A%22Innan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Diddahally%20R.%22%2C%22lastName%22%3A%22Govindaraju%22%7D%5D%2C%22abstractNote%22%3A%22Mutation%20accumulation%20has%20been%20proposed%20as%20a%20cause%20of%20senescence.%20During%20this%20process%2C%20age-related%20genetic%20and%20epigenetic%20mutations%20steadily%20accumulate.%20Cascading%20deleterious%20effects%20of%20mutations%20might%20initiate%20a%20steady%20%5Cu201caccumulation%20of%20deficits%5Cu201d%20in%20cells%2C%20despite%20the%20existence%20of%20repair%20mechanisms%2C%20leading%20to%20cellular%20senescence%20and%20functional%20decline%20of%20tissues%20and%20organs%2C%20which%20ultimately%20manifest%20as%20frailty%20and%20disease.%20Here%2C%20we%20investigate%20several%20of%20these%20aspects%20in%20differentiating%20cell%20populations%20through%20modeling%20and%20simulation%20using%20the%20Moran%20birth%5Cu2013death%20%28demographic%29%20process%2C%20under%20several%20scenarios%20of%20mutation%20accumulation.%20Deleterious%20mutations%20seem%20to%20rapidly%20accumulate%20particularly%20early%20in%20the%20course%20of%20life%2C%20during%20which%20the%20rate%20of%20cell%20division%20is%20high%2C%20thereby%20exerting%20a%20greater%20effect%20on%20subsequent%20cellular%20senescence.%20Our%20results%20are%20compatible%20with%20the%20principle%20of%20the%20Muller%5Cu2019s%20ratchet%20taking%20place%20in%20asexually%20reproducing%20organisms.%20The%20ratchet%20speed%20in%20a%20given%20tissue%20depends%20on%20the%20size%20of%20the%20cell%20population%2C%20mutation%20rate%20and%20the%20impact%20of%20such%20mutations%20on%20cell%20phenotypes.%20It%20varies%20substantially%20among%20cells%20in%20different%20tissues%20and%20organs%20due%20to%20heterogeneity%20in%20relation%20to%20cell%20and%20organ-specific%20demographic%20features.%20Ratchet%20accelerates%20particularly%20after%20middle%20age%2C%20resulting%20in%20a%20synergistic%20fitness%20decay%20at%20the%20level%20of%20cell%20populations.%20We%20extend%20Fisher%5Cu2019s%20average%20excess%20concept%20and%20rank%20order%20scale%20to%20interpret%20differential%20phenotypic%20effects%20of%20the%20increase%20of%20the%20mutation%20load%20among%20cell%20populations%20within%20a%20given%20tissue.%20We%20postulate%20that%20classical%20evolutionary%20genetic%20models%20can%20explain%2C%20at%20least%20in%20part%2C%20the%20origins%20of%20frailty%2C%20subclinical%20conditions%2C%20morbidity%20and%20the%20health%20consequences%20of%20senescence.%22%2C%22date%22%3A%222020-03-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00439-019-02067-9%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-019-02067-9%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221432-1203%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-06-07T14%3A47%3A32Z%22%7D%7D%2C%7B%22key%22%3A%22MWNZHV3W%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Benayoun%20and%20Veitia%22%2C%22parsedDate%22%3A%222020-03%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBenayoun%2C%20B.%20A.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282020%29.%20Special%20issue%20on%20%26%23x201C%3BMolecular%20genetics%20of%20aging%20and%20longevity%26%23x201D%3B%3A%20a%20critical%20time%20in%20the%20field%20of%20geroscience.%20%26lt%3Bi%26gt%3BHuman%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B139%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20275%26%23×2013%3B276.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-020-02125-7%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-020-02125-7%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Special%20issue%20on%20%5C%22Molecular%20genetics%20of%20aging%20and%20longevity%5C%22%3A%20a%20critical%20time%20in%20the%20field%20of%20geroscience%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B%5Cu00e9r%5Cu00e9nice%20A.%22%2C%22lastName%22%3A%22Benayoun%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222020-03%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00439-020-02125-7%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221432-1203%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22QKLWM6BT%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Grassmann%20et%20al.%22%2C%22parsedDate%22%3A%222020-03%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BGrassmann%2C%20F.%2C%20International%20AMD%20Genomics%20Consortium%20%28IAMDGC%29%2C%20Weber%2C%20B.%20H.%20F.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282020%29.%20Insights%20into%20the%20loss%20of%20the%20Y%20chromosome%20with%20age%20in%20control%20individuals%20and%20in%20patients%20with%20age-related%20macular%20degeneration%20using%20genotyping%20microarray%20data.%20%26lt%3Bi%26gt%3BHuman%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B139%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20401%26%23×2013%3B407.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-019-02029-1%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00439-019-02029-1%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Insights%20into%20the%20loss%20of%20the%20Y%20chromosome%20with%20age%20in%20control%20individuals%20and%20in%20patients%20with%20age-related%20macular%20degeneration%20using%20genotyping%20microarray%20data%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Felix%22%2C%22lastName%22%3A%22Grassmann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22name%22%3A%22International%20AMD%20Genomics%20Consortium%20%28IAMDGC%29%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bernhard%20H.%20F.%22%2C%22lastName%22%3A%22Weber%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22The%20extent%20of%20aneuploidy%20of%20the%20sex%20chromosomes%20increases%20with%20age%20in%20human%20leukocytes.%20Here%2C%20we%20re-explore%20the%20dynamics%20of%20normal%20loss%20of%20the%20Y%20chromosome%20%28LOY%29%20with%20age%20based%20on%20microarray%20data%20using%20two%20exponential%20models%20and%20two%20different%20ways%20to%20estimate%20the%20fraction%20of%20LOY.%20This%20analysis%20shows%20the%20existence%20of%20a%20significant%20correlation%20between%20the%20fraction%20of%20LOY%20estimated%20from%20molecular%20cytogenetics%20and%20genotyping%20microarray%20data.%20Although%20the%20specific%20estimates%20of%20the%20parameters%20for%20the%20two%20exponential%20models%20are%20different%20from%20those%20derived%20from%20cytogenetics%20data%2C%20the%20present%20analysis%20in%20an%20independent%20dataset%20of%20normal%20individuals%20confirms%20that%20X0%20cells%20have%20a%20selective%20advantage%20over%20XY%20cells.%20Moreover%2C%20patients%20with%20age-related%20macular%20degeneration%20display%20higher%20fraction%20of%20LOY%20values%20and%20seem%20to%20have%20a%20predisposition%20to%20lose%20their%20Y%20chromosome%20even%20at%20young%20ages%20compared%20to%20control%20individuals.%20As%20there%20are%20no%20data%20available%20for%20the%20same%20individuals%20at%20different%20time%20points%2C%20the%20parameters%20reported%20here%20are%20average%20values%20drawn%20from%20population%20analyses.%22%2C%22date%22%3A%222020-03%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00439-019-02029-1%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221432-1203%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%22NNWFWZW5%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Penrad-Mobayed%20et%20al.%22%2C%22parsedDate%22%3A%222020-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPenrad-Mobayed%2C%20M.%2C%20Perrin%2C%20C.%2C%20Herman%2C%20L.%2C%20Todeschini%2C%20A.-L.%2C%20Nigon%2C%20F.%2C%20Cosson%2C%20B.%2C%20Caburet%2C%20S.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282020%29.%20Conventional%20and%20unconventional%20interactions%20of%20the%20transcription%20factor%20FOXL2%20uncovered%20by%20a%20proteome-wide%20analysis.%20%26lt%3Bi%26gt%3BFASEB%20Journal%3A%20Official%20Publication%20of%20the%20Federation%20of%20American%20Societies%20for%20Experimental%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B34%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%20571%26%23×2013%3B587.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1096%5C%2Ffj.201901573R%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1096%5C%2Ffj.201901573R%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Conventional%20and%20unconventional%20interactions%20of%20the%20transcription%20factor%20FOXL2%20uncovered%20by%20a%20proteome-wide%20analysis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22May%22%2C%22lastName%22%3A%22Penrad-Mobayed%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Caroline%22%2C%22lastName%22%3A%22Perrin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laetitia%22%2C%22lastName%22%3A%22Herman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fabienne%22%2C%22lastName%22%3A%22Nigon%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bertrand%22%2C%22lastName%22%3A%22Cosson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Caburet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Beyond%20the%20study%20of%20its%20transcriptional%20target%20genes%2C%20the%20identification%20of%20the%20various%20interactors%20of%20a%20transcription%20factor%20%28TF%29%20is%20crucial%20to%20understand%20its%20diverse%20cellular%20roles.%20We%20focused%20on%20FOXL2%2C%20a%20winged-helix%20forkhead%20TF%20important%20for%20ovarian%20development%20and%20maintenance.%20FOXL2%20has%20been%20implicated%20in%20diverse%20cellular%20processes%2C%20including%20apoptosis%2C%20the%20control%20of%20cell%20cycle%20or%20the%20regulation%20of%20steroid%20hormone%20synthesis.%20To%20reliably%20identify%20partners%20of%20endogenous%20FOXL2%2C%20we%20performed%20a%20proteome-wide%20analysis%20using%20co-immunoprecipitation%20in%20the%20murine%20granulosa%20cell-derived%20AT29c%20and%20the%20pituitary-derived%20alpha-T3%20cell%20lines%2C%20using%20three%20antibodies%20targeting%20different%20parts%20of%20the%20protein.%20Following%20a%20stringent%20selection%20of%20mass%20spectrometry%20data%20on%20the%20basis%20of%20identification%20reliability%20and%20protein%20enrichment%2C%20we%20identified%20a%20core%20set%20of%20255%20partners%20common%20to%20both%20cell%20lines.%20Their%20analysis%20showed%20that%20we%20could%20co-precipitate%20several%20complexes%20involved%20in%20mRNA%20processing%2C%20chromatin%20remodeling%20and%20DNA%20replication%20and%20repair.%20We%20further%20validated%20%28direct%20and%5C%2For%20indirect%29%20interactions%20with%20selected%20partners%2C%20suggesting%20an%20unexpected%20role%20for%20FOXL2%20in%20those%20processes.%20Overall%2C%20this%20comprehensive%20analysis%20of%20the%20endogenous%20FOXL2%20interactome%20sheds%20light%20on%20its%20numerous%20and%20diverse%20interactors%20and%20unconventional%20cellular%20roles.%22%2C%22date%22%3A%222020-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1096%5C%2Ffj.201901573R%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221530-6860%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A07Z%22%7D%7D%2C%7B%22key%22%3A%2273TJCUT2%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Shi%20et%20al.%22%2C%22parsedDate%22%3A%222020%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BShi%2C%20X.%2C%20Chen%2C%20C.%2C%20Yang%2C%20H.%2C%20Hou%2C%20J.%2C%20Ji%2C%20T.%2C%20Cheng%2C%20J.%2C%20Veitia%2C%20R.%20A.%2C%20%26amp%3B%20Birchler%2C%20J.%20A.%20%282020%29.%20The%20Gene%20Balance%20Hypothesis%3A%20Epigenetics%20and%20Dosage%20Effects%20in%20Plants.%20%26lt%3Bi%26gt%3BMethods%20in%20Molecular%20Biology%20%28Clifton%2C%20N.J.%29%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B2093%26lt%3B%5C%2Fi%26gt%3B%2C%20161%26%23×2013%3B171.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-1-0716-0179-2_12%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-1-0716-0179-2_12%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20Gene%20Balance%20Hypothesis%3A%20Epigenetics%20and%20Dosage%20Effects%20in%20Plants%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xiaowen%22%2C%22lastName%22%3A%22Shi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chen%22%2C%22lastName%22%3A%22Chen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hua%22%2C%22lastName%22%3A%22Yang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jie%22%2C%22lastName%22%3A%22Hou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tieming%22%2C%22lastName%22%3A%22Ji%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jianlin%22%2C%22lastName%22%3A%22Cheng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%20A.%22%2C%22lastName%22%3A%22Birchler%22%7D%5D%2C%22abstractNote%22%3A%22Dosage%20effects%20in%20plants%20are%20caused%20by%20changes%20in%20the%20copy%20number%20of%20chromosomes%2C%20segments%20of%20chromosomes%2C%20or%20multiples%20of%20individual%20genes.%20Genes%20often%20exhibit%20a%20dosage%20effect%20in%20which%20the%20amount%20of%20product%20is%20closely%20correlated%20with%20the%20number%20of%20copies%20present.%20However%2C%20when%20larger%20segments%20of%20chromosomes%20are%20varied%2C%20there%20are%20trans-acting%20effects%20across%20the%20genome%20that%20are%20unleashed%20that%20modulate%20gene%20expression%20in%20cascading%20effects.%20These%20appear%20to%20be%20mediated%20by%20the%20stoichiometric%20relationship%20of%20gene%20regulatory%20machineries.%20There%20are%20both%20positive%20and%20negative%20modulations%20of%20target%20gene%20expression%2C%20but%20the%20latter%20is%20the%20plurality%20effect.%20When%20this%20inverse%20effect%20is%20combined%20with%20a%20dosage%20effect%2C%20compensation%20for%20a%20gene%20can%20occur%20in%20which%20its%20expression%20is%20similar%20to%20the%20normal%20diploid%20regardless%20of%20the%20change%20in%20chromosomal%20dosage.%20In%20contrast%2C%20changing%20the%20whole%20genome%20in%20a%20polyploidy%20series%20has%20fewer%20relative%20effects%20as%20the%20stoichiometric%20relationship%20is%20not%20disrupted.%20Together%2C%20these%20observations%20suggest%20that%20the%20stoichiometry%20of%20gene%20regulation%20is%20important%20as%20a%20reflection%20of%20the%20mode%20of%20assembly%20of%20the%20individual%20subunits%20involved%20in%20the%20effective%20regulatory%20macromolecular%20complexes.%20This%20principle%20has%20implications%20for%20gene%20expression%20mechanisms%2C%20quantitative%20trait%20genetics%2C%20and%20the%20evolution%20of%20genes%20depending%20on%20the%20mode%20of%20duplication%2C%20either%20segmentally%20or%20via%20whole-genome%20duplication.%22%2C%22date%22%3A%222020%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1007%5C%2F978-1-0716-0179-2_12%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221940-6029%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22TWVG9ACD%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222019-11%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282019%29.%20MIRAGE%20Syndrome%3A%20Phenotypic%20Rescue%20by%20Somatic%20Mutation%20and%20Selection.%20%26lt%3Bi%26gt%3BTrends%20in%20Molecular%20Medicine%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B25%26lt%3B%5C%2Fi%26gt%3B%2811%29%2C%20937%26%23×2013%3B940.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.molmed.2019.08.008%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.molmed.2019.08.008%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22MIRAGE%20Syndrome%3A%20Phenotypic%20Rescue%20by%20Somatic%20Mutation%20and%20Selection%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22MIRAGE%20syndrome%2C%20a%20multisystem%20disorder%2C%20results%20from%20heterozygous%20gain-of-function%20mutations%20in%20SAMD9%2C%20which%20encodes%20a%20growth%20suppressor%2C%20located%20on%20chromosome%207.%20Somatic%20changes%20involving%20loss-of-function%20mutations%20of%20the%20altered%20SAMD9%20allele%20or%20loss%20of%20chromosome%207%20act%20as%20phenotypic%20modifiers%2C%20providing%20a%20typical%20example%20of%20somatic%20mutation%20and%20selection%20process.%22%2C%22date%22%3A%222019-11%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.molmed.2019.08.008%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221471-499X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A07Z%22%7D%7D%2C%7B%22key%22%3A%22GQ6KKM2Y%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Johnson%20et%20al.%22%2C%22parsedDate%22%3A%222019-10%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BJohnson%2C%20A.%20F.%2C%20Nguyen%2C%20H.%20T.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282019%29.%20Causes%20and%20effects%20of%20haploinsufficiency.%20%26lt%3Bi%26gt%3BBiological%20Reviews%20of%20the%20Cambridge%20Philosophical%20Society%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B94%26lt%3B%5C%2Fi%26gt%3B%285%29%2C%201774%26%23×2013%3B1785.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fbrv.12527%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fbrv.12527%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Causes%20and%20effects%20of%20haploinsufficiency%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Adam%20F.%22%2C%22lastName%22%3A%22Johnson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ha%20T.%22%2C%22lastName%22%3A%22Nguyen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Haploinsufficiency%20is%20a%20form%20of%20genetic%20dominance%20and%20is%20the%20underlying%20mechanism%20of%20numerous%20human%20inherited%20conditions%20in%20which%20the%20causal%20genes%20are%20sensitive%20to%20altered%20dosage.%20This%20review%20examines%20the%20poorly%20understood%20relationships%20between%20haploinsufficiency%2C%20dosage%20sensitivity%20and%20genetic%20dominance%2C%20whose%20common%20theme%20is%20the%20existence%20of%20nonlinear%20relationships%20between%20genotype%20and%20phenotype.%20We%20present%20an%20up-to-date%20account%20of%20the%20bases%20of%20haploinsufficiency%20from%20the%20perspective%20of%20theoretical%20and%20experimental%20models.%20We%20also%20discuss%20human%20conditions%20caused%20by%20haploinsufficiency%2C%20including%20developmental%20syndromes%20and%20cancer.%20Connections%20between%20the%20understanding%20of%20these%20conditions%26%23039%3B%20genetic%20mechanisms%20and%20advances%20in%20treatments%20are%20also%20described.%22%2C%22date%22%3A%222019-10%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fbrv.12527%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221469-185X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%222SLAJ6NR%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222019-09-19%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282019%29.%20AFF3%3A%20a%20new%20player%20in%20maintaining%20XIST%20monoallelic%20expression.%20%26lt%3Bi%26gt%3BJournal%20of%20Molecular%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B11%26lt%3B%5C%2Fi%26gt%3B%289%29%2C%20723%26%23×2013%3B724.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjmcb%5C%2Fmjy082%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjmcb%5C%2Fmjy082%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22AFF3%3A%20a%20new%20player%20in%20maintaining%20XIST%20monoallelic%20expression%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222019-09-19%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fjmcb%5C%2Fmjy082%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221759-4685%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%226PF8DKAD%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222019-08-19%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282019%29.%20Darwinian%20selection%20within%20an%20individual%20or%20somatic%20selection%3A%20facts%20and%20models.%20%26lt%3Bi%26gt%3BJournal%20of%20Molecular%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B11%26lt%3B%5C%2Fi%26gt%3B%288%29%2C%20719%26%23×2013%3B722.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjmcb%5C%2Fmjz014%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjmcb%5C%2Fmjz014%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Darwinian%20selection%20within%20an%20individual%20or%20somatic%20selection%3A%20facts%20and%20models%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222019-08-19%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fjmcb%5C%2Fmjz014%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221759-4685%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%22IB7T7MU6%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222019-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282019%29.%20DNA%20Content%2C%20Cell%20Size%2C%20and%20Cell%20Senescence.%20%26lt%3Bi%26gt%3BTrends%20in%20Biochemical%20Sciences%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B44%26lt%3B%5C%2Fi%26gt%3B%288%29%2C%20645%26%23×2013%3B647.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tibs.2019.04.013%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tibs.2019.04.013%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22DNA%20Content%2C%20Cell%20Size%2C%20and%20Cell%20Senescence%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22A%20recent%20study%20assessed%20the%20impact%20of%20cell%20size%20on%20various%20cell%20properties.%20Oversized%20yeast%20cells%20display%20slow%20cell%20division%2C%20cytoplasmic%20dilution%2C%20and%20transcriptomic%20and%20proteomic%20alterations.%20It%20highlights%20commonalities%20between%20aging%20yeast%20and%20mammalian%20cells%2C%20suggesting%20the%20existence%20of%20a%20range%20of%20DNA%20content%3A%20cell%20volume%20ratio%20ensuring%20optimal%20function.%22%2C%22date%22%3A%222019-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tibs.2019.04.013%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220968-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%22X3SP24YS%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Elzaiat%20et%20al.%22%2C%22parsedDate%22%3A%222019-07%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BElzaiat%2C%20M.%2C%20Herman%2C%20L.%2C%20Legois%2C%20B.%2C%20L%26%23xE9%3Bger%2C%20T.%2C%20Todeschini%2C%20A.-L.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282019%29.%20High-throughput%20Exploration%20of%20the%20Network%20Dependent%20on%20AKT1%20in%20Mouse%20Ovarian%20Granulosa%20Cells.%20%26lt%3Bi%26gt%3BMolecular%20%26amp%3B%20Cellular%20Proteomics%3A%20MCP%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B18%26lt%3B%5C%2Fi%26gt%3B%287%29%2C%201307%26%23×2013%3B1319.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1074%5C%2Fmcp.RA119.0014613%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1074%5C%2Fmcp.RA119.0014613%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22High-throughput%20Exploration%20of%20the%20Network%20Dependent%20on%20AKT1%20in%20Mouse%20Ovarian%20Granulosa%20Cells%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ma%5Cu00ebva%22%2C%22lastName%22%3A%22Elzaiat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laetitia%22%2C%22lastName%22%3A%22Herman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B%5Cu00e9rang%5Cu00e8re%22%2C%22lastName%22%3A%22Legois%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thibaut%22%2C%22lastName%22%3A%22L%5Cu00e9ger%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22The%20PI3K%5C%2FAKT%20signaling%20pathway%20is%20known%20to%20regulate%20a%20broad%20range%20of%20cellular%20processes%2C%20and%20it%20is%20often%20altered%20in%20several%20types%20of%20cancers.%20Recently%2C%20somatic%20AKT1%20mutations%20leading%20to%20a%20strong%20activation%20of%20this%20kinase%20have%20been%20reported%20in%20juvenile%20granulosa%20cell%20tumors.%20However%2C%20the%20molecular%20role%20of%20AKT1%20in%20the%20supporting%20cell%20lineage%20of%20the%20ovary%20is%20still%20poorly%20understood.%20To%20get%20insights%20into%20its%20function%20in%20such%20cells%2C%20we%20depleted%20Akt1%20in%20murine%20primary%20granulosa%20cells%20and%20assessed%20the%20molecular%20consequences%20at%20both%20the%20transcript%20and%20protein%20levels.%20We%20were%20able%20to%20corroborate%20the%20involvement%20of%20AKT1%20in%20the%20regulation%20of%20metabolism%2C%20apoptosis%2C%20cell%20cycle%2C%20or%20cytoskeleton%20dynamics%20in%20this%20ovarian%20cell%20type.%20Consistently%2C%20we%20showed%20in%20established%20granulosa%20cells%20that%20depletion%20of%20Akt1%20provoked%20altered%20directional%20persistent%20migration%20and%20increased%20its%20velocity.%20This%20study%20also%20allowed%20us%20to%20put%20forward%20new%20direct%20and%20indirect%20targets%20of%20the%20kinase.%20Indeed%2C%20a%20series%20of%20proteins%20involved%20in%20intracellular%20transport%20and%20mitochondrial%20physiology%20were%20significantly%20affected%20by%20Akt1%20depletion.%20Using%20in%20silico%20analyses%2C%20we%20also%20propose%20a%20set%20of%20kinases%20and%20transcription%20factors%20that%20can%20mediate%20the%20action%20of%20AKT1%20on%20the%20deregulated%20transcripts%20and%20proteins.%20Taken%20altogether%2C%20our%20results%20provide%20a%20resource%20of%20direct%20and%20indirect%20AKT1%20targets%20in%20granulosa%20cells%20and%20may%20help%20understand%20its%20roles%20in%20this%20ovarian%20cell%20type.%22%2C%22date%22%3A%222019-07%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1074%5C%2Fmcp.RA119.0014613%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221535-9484%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%222URIF8MD%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222019-04%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282019%29.%20Further%20quantitative%20insights%20into%20the%20decrease%20of%20heteroplasmy%20of%20m.3243A%26gt%3BG%20with%20age%20in%20leukocytes.%20%26lt%3Bi%26gt%3BClinical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B95%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20542%26%23×2013%3B543.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13496%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13496%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Further%20quantitative%20insights%20into%20the%20decrease%20of%20heteroplasmy%20of%20m.3243A%3EG%20with%20age%20in%20leukocytes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222019-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fcge.13496%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221399-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%22FTLUFKHU%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Caburet%20et%20al.%22%2C%22parsedDate%22%3A%222019-04%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BCaburet%2C%20S.%2C%20Todeschini%2C%20A.-L.%2C%20Petrillo%2C%20C.%2C%20Martini%2C%20E.%2C%20Farran%2C%20N.%20D.%2C%20Legois%2C%20B.%2C%20Livera%2C%20G.%2C%20Younis%2C%20J.%20S.%2C%20Shalev%2C%20S.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282019%29.%20A%20truncating%20MEIOB%20mutation%20responsible%20for%20familial%20primary%20ovarian%20insufficiency%20abolishes%20its%20interaction%20with%20its%20partner%20SPATA22%20and%20their%20recruitment%20to%20DNA%20double-strand%20breaks.%20%26lt%3Bi%26gt%3BEBioMedicine%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B42%26lt%3B%5C%2Fi%26gt%3B%2C%20524%26%23×2013%3B531.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.ebiom.2019.03.075%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.ebiom.2019.03.075%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20truncating%20MEIOB%20mutation%20responsible%20for%20familial%20primary%20ovarian%20insufficiency%20abolishes%20its%20interaction%20with%20its%20partner%20SPATA22%20and%20their%20recruitment%20to%20DNA%20double-strand%20breaks%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Caburet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Laure%22%2C%22lastName%22%3A%22Todeschini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cynthia%22%2C%22lastName%22%3A%22Petrillo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Emmanuelle%22%2C%22lastName%22%3A%22Martini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nada%20D.%22%2C%22lastName%22%3A%22Farran%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B%5Cu00e9rang%5Cu00e8re%22%2C%22lastName%22%3A%22Legois%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gabriel%22%2C%22lastName%22%3A%22Livera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Johnny%20S.%22%2C%22lastName%22%3A%22Younis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stavit%22%2C%22lastName%22%3A%22Shalev%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22BACKGROUND%3A%20Primary%20Ovarian%20Insufficiency%20%28POI%29%2C%20a%20major%20cause%20of%20infertility%2C%20affects%20about%201-3%25%20of%20women%20under%20forty%20years%20of%20age.%20Although%20there%20is%20a%20growing%20list%20of%20causal%20genetic%20alterations%2C%20POI%20remains%20mostly%20idiopathic.%5CnMETHODS%3A%20We%20performed%20exome%20sequencing%20%28WES%29%20of%20two%20sisters%20affected%20with%20POI%2C%20one%20unaffected%20sister%20and%20their%20mother%20from%20a%20consanguineous%20family.%20We%20assessed%20the%20impact%20of%20the%20identified%20MEIOB%20variant%20with%20a%20minigene%20assay%20and%20by%20sequencing%20illegitimate%20transcripts%20from%20the%20proband%26%23039%3Bs%20leukocytes.%20We%20studied%20its%20functional%20impact%20on%20the%20interaction%20between%20MEIOB%20with%20its%20partner%20SPATA22%20and%20their%20localization%20to%20DNA%20double-strand%20breaks%20%28DSB%29.%5CnFINDINGS%3A%20We%20identified%20a%20homozygous%20variant%20in%20the%20last%20base%20of%20exon%2012%20of%20MEIOB%2C%20which%20encodes%20a%20factor%20essential%20for%20meiotic%20recombination.%20This%20variant%20was%20predicted%20to%20strongly%20affect%20MEIOB%20pre-mRNA%20splicing.%20Consistently%2C%20a%20minigene%20assay%20showed%20that%20the%20variant%20induced%20exon%2012%20skipping%2C%20which%20was%20confirmed%20in%20vivo%20in%20the%20proband%26%23039%3Bs%20leukocytes.%20Aberrant%20splicing%20leads%20to%20the%20production%20of%20a%20C-terminally%20truncated%20protein%20that%20cannot%20interact%20with%20SPATA22%2C%20abolishing%20their%20recruitment%20to%20DSBs.%5CnINTERPRETATION%3A%20This%20truncating%20MEIOB%20variant%20is%20expected%20to%20provoke%20meiotic%20defects%20and%20a%20depleted%20follicular%20stock%2C%20as%20in%20Meiob-%5C%2F-%20mice.%20This%20is%20the%20first%20molecular%20defect%20reported%20in%20a%20meiosis-specific%20single-stranded%20DNA-binding%20protein%20%28SSB%29%20responsible%20for%20POI.%20We%20hypothesise%20that%20alterations%20in%20other%20SSB%20proteins%20could%20explain%20cases%20of%20syndromic%20or%20isolated%20ovarian%20insufficiency.%20FUND%3A%20Universit%5Cu00e9%20Paris%20Diderot%2C%20Fondation%20pour%20la%20Recherche%20M%5Cu00e9dicale%2C%20Fondation%20ARC%20contre%20le%20cancer%2C%20Commissariat%20%5Cu00e0%20l%26%23039%3BEnergie%20Atomique%20and%20Institut%20Universitaire%20de%20France.%22%2C%22date%22%3A%222019-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.ebiom.2019.03.075%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222352-3964%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%22UT8H6QX5%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Ouimette%20et%20al.%22%2C%22parsedDate%22%3A%222019-02%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BOuimette%2C%20J.-F.%2C%20Rougeulle%2C%20C.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282019%29.%20Three-dimensional%20genome%20architecture%20in%20health%20and%20disease.%20%26lt%3Bi%26gt%3BClinical%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B95%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20189%26%23×2013%3B198.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13219%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1111%5C%2Fcge.13219%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Three-dimensional%20genome%20architecture%20in%20health%20and%20disease%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J.-F.%22%2C%22lastName%22%3A%22Ouimette%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C.%22%2C%22lastName%22%3A%22Rougeulle%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22More%20than%20a%20decade%20of%20massive%20DNA%20sequencing%20efforts%20have%20generated%20a%20large%20body%20of%20genomic%2C%20transcriptomic%20and%20epigenomic%20information%20that%20has%20provided%20a%20more%20and%20more%20detailed%20view%20of%20the%20functional%20elements%20and%20transactions%20within%20the%20human%20genome.%20Considerable%20efforts%20have%20also%20focused%20on%20linking%20these%20elements%20with%20one%20another%20by%20mapping%20their%20interactions%20and%20by%20establishing%203-dimensional%20%283D%29%20genomic%20landscapes%20in%20various%20cell%20and%20tissue%20types.%20In%20parallel%2C%20multiple%20studies%20have%20associated%20genomic%20deletions%2C%20duplications%20and%20other%20rearrangements%20with%20human%20pathologies.%20In%20this%20review%2C%20we%20explore%20recent%20progresses%20that%20have%20allowed%20connecting%20disease-causing%20alterations%20with%20perturbations%20of%20the%203D%20genome%20organization.%22%2C%22date%22%3A%222019-02%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1111%5C%2Fcge.13219%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221399-0004%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%22A8RJ66VG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Birchler%20and%20Veitia%22%2C%22parsedDate%22%3A%222019%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBirchler%2C%20J.%20A.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282019%29.%20Genomic%20Balance%20and%20Speciation.%20%26lt%3Bi%26gt%3BEpigenetics%20Insights%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B12%26lt%3B%5C%2Fi%26gt%3B%2C%202516865719840291.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1177%5C%2F2516865719840291%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1177%5C%2F2516865719840291%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Genomic%20Balance%20and%20Speciation%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%20A.%22%2C%22lastName%22%3A%22Birchler%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22The%20role%20of%20genomic%20balance%20in%20accumulating%20species%20hybrid%20incompatibilities%20is%20discussed.%20Aneuploidy%20has%20been%20shown%20to%20produce%20more%20global%20modulations%20than%20polyploidy%20with%20the%20responsible%20genes%20being%20transcription%20factors%20and%20signaling%20components%20involved%20in%20molecular%20complexes%2C%20illustrating%20a%20stoichiometric%20component%20to%20gene%20expression.%20Genomic%20imbalance%20is%20usually%20detrimental%20to%20the%20organism%20and%20in%20many%20cases%20results%20in%20lethality.%20Here%2C%20it%20is%20proposed%20that%20once%20gene%20flow%20is%20prevented%20between%20or%20within%20populations%20by%20various%20speciation%20initiating%20processes%2C%20the%20stoichiometric%20relationship%20of%20members%20of%20macromolecular%20complexes%20can%20change%20via%20compensatory%20drift%20with%20the%20eventual%20result%20of%20newly%20established%20functional%20balances.%20However%2C%20when%20these%20new%20relationships%20are%20brought%20together%20in%20interspecific%20hybrids%2C%20detrimental%20consequences%20will%20occur.%20We%20suggest%20that%20these%20detrimental%20interactions%20contribute%20to%20hybrid%20incompatibilities.%22%2C%22date%22%3A%222019%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1177%5C%2F2516865719840291%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222516-8657%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A23Z%22%7D%7D%2C%7B%22key%22%3A%22YBYDTGYB%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222018-12%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282018%29.%20On%20the%20loss%20of%20human%20sex%20chromosomes%20in%20lymphocytes%20with%20age%3A%20a%20quantitative%20treatment.%20%26lt%3Bi%26gt%3BEuropean%20Journal%20of%20Human%20Genetics%3A%20EJHG%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B26%26lt%3B%5C%2Fi%26gt%3B%2812%29%2C%201875%26%23×2013%3B1878.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41431-018-0225-0%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41431-018-0225-0%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22On%20the%20loss%20of%20human%20sex%20chromosomes%20in%20lymphocytes%20with%20age%3A%20a%20quantitative%20treatment%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Increasing%20levels%20of%20aneuploidy%20of%20the%20sex%20chromosomes%20in%20human%20lymphocytes%20with%20age%20have%20been%20noted%20for%20several%20decades.%20The%20percentage%20of%20chromosome%20Y%20loss%20can%20reach%20up%20to%201.5%25%20or%20even%20more%2C%20whereas%20the%20levels%20of%20X0%20cells%20in%20females%20can%20increase%20up%20to%205%25%20with%20age.%20Here%2C%20I%20propose%20simple%20mathematical%20models%20of%20the%20dynamics%20of%20%26%23039%3Bnormal%26%23039%3B%20sex%20chromosome%20loss%20with%20age.%20These%20exponential%20models%20provide%20more%20mechanistic%20insights%20than%20linear%20regressions.%20They%20account%20for%20the%20lower%20incidence%20of%20sex%20chromosome%20loss%20in%20young%20individuals%20and%20its%20increase%20with%20age.%20Moreover%2C%20the%20exponential%20models%20show%20that%20aneuploidy%20of%20the%20sex%20chromosomes%20provides%20a%20selective%20advantage.%20As%20there%20are%20no%20longitudinal%20data%20available%20%28for%20the%20same%20individual%20at%20different%20time%20points%29%2C%20the%20parameters%20reported%20here%20are%20average%20values%20derived%20from%20a%20population.%20Hopefully%2C%20this%20study%20will%20stimulate%20further%20work%20based%20on%20next-generation%20technologies%20to%20obtain%20better%20estimates%20of%20sex%20chromosome%20aneuploidy%20and%20of%20the%20parameters%20of%20the%20models%20discussed%20here.%22%2C%22date%22%3A%222018-12%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41431-018-0225-0%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221476-5438%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%225N6NK7L5%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Veitia%22%2C%22parsedDate%22%3A%222018-06%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVeitia%2C%20R.%20A.%20%282018%29.%20Dosage%20effects%20in%20morphogenetic%20gradients%20of%20transcription%20factors%3A%20insights%20from%20a%20simple%20mathematical%20model.%20%26lt%3Bi%26gt%3BJournal%20of%20Genetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B97%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20365%26%23×2013%3B370.%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Dosage%20effects%20in%20morphogenetic%20gradients%20of%20transcription%20factors%3A%20insights%20from%20a%20simple%20mathematical%20model%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22The%20classical%20Hill%20equation%20is%20the%20simplest%20way%20to%20model%20sharp%20transcriptional%20responses%20%28TR%29%20to%20changing%20concentrations%20of%20a%20regulator.%20Such%20steep%20sigmoidal%20transitions%20of%20the%20TR%20are%20often%20involved%20in%20the%20creation%20of%20boundaries%20or%20domains%20during%20development.%20Here%2C%20I%20use%20the%20Hill%20function%20to%20explore%20the%20response%20of%20a%20promoter%20to%20a%20gradient%20of%20a%20transcription%20factor%20%28TF%29.%20Specifically%2C%20I%20examine%20how%20various%20sources%20of%20nonlinearity%20%28such%20as%20that%20imposed%20by%20the%20number%20of%20TF-binding%20sites%29%20and%20the%20nonlinearity%20of%20the%20gradient%20itself%20influence%20TR%20and%20modulate%20its%20robustness%20to%20variations%20in%20the%20dosage%20of%20morphogen.%20This%20qualitative%20analysis%20underscores%20how%20nonlinearities%20can%20modulate%20the%20robustness%20of%20TR%20and%20is%20a%20convenient%20way%20to%20convey%20concepts%20in%20a%20teaching%20setting.%22%2C%22date%22%3A%222018-06%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220973-7731%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%22BIP65NKN%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Huhtaniemi%20et%20al.%22%2C%22parsedDate%22%3A%222018-06%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BHuhtaniemi%2C%20I.%2C%20Hovatta%2C%20O.%2C%20La%20Marca%2C%20A.%2C%20Livera%2C%20G.%2C%20Monniaux%2C%20D.%2C%20Persani%2C%20L.%2C%20Heddar%2C%20A.%2C%20Jarzabek%2C%20K.%2C%20Laisk-Podar%2C%20T.%2C%20Salumets%2C%20A.%2C%20Tapanainen%2C%20J.%20S.%2C%20Veitia%2C%20R.%20A.%2C%20Visser%2C%20J.%20A.%2C%20Wieacker%2C%20P.%2C%20Wolczynski%2C%20S.%2C%20%26amp%3B%20Misrahi%2C%20M.%20%282018%29.%20Advances%20in%20the%20Molecular%20Pathophysiology%2C%20Genetics%2C%20and%20Treatment%20of%20Primary%20Ovarian%20Insufficiency.%20%26lt%3Bi%26gt%3BTrends%20in%20Endocrinology%20and%20Metabolism%3A%20TEM%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B29%26lt%3B%5C%2Fi%26gt%3B%286%29%2C%20400%26%23×2013%3B419.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tem.2018.03.010%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tem.2018.03.010%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Advances%20in%20the%20Molecular%20Pathophysiology%2C%20Genetics%2C%20and%20Treatment%20of%20Primary%20Ovarian%20Insufficiency%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ilpo%22%2C%22lastName%22%3A%22Huhtaniemi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Outi%22%2C%22lastName%22%3A%22Hovatta%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Antonio%22%2C%22lastName%22%3A%22La%20Marca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gabriel%22%2C%22lastName%22%3A%22Livera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Danielle%22%2C%22lastName%22%3A%22Monniaux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Luca%22%2C%22lastName%22%3A%22Persani%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Abdelkader%22%2C%22lastName%22%3A%22Heddar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Katarzyna%22%2C%22lastName%22%3A%22Jarzabek%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Triin%22%2C%22lastName%22%3A%22Laisk-Podar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andres%22%2C%22lastName%22%3A%22Salumets%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juha%20S.%22%2C%22lastName%22%3A%22Tapanainen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Reiner%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jenny%20A.%22%2C%22lastName%22%3A%22Visser%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%22%2C%22lastName%22%3A%22Wieacker%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Slawomir%22%2C%22lastName%22%3A%22Wolczynski%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Micheline%22%2C%22lastName%22%3A%22Misrahi%22%7D%5D%2C%22abstractNote%22%3A%22Primary%20ovarian%20insufficiency%20%28POI%29%20affects%20%5Cu223c1%25%20of%20women%20before%2040%20years%20of%20age.%20The%20recent%20leap%20in%20genetic%20knowledge%20obtained%20by%20next%20generation%20sequencing%20%28NGS%29%20together%20with%20animal%20models%20has%20further%20elucidated%20its%20molecular%20pathogenesis%2C%20identifying%20novel%20genes%5C%2Fpathways.%20Mutations%20of%20%26gt%3B60%20genes%20emphasize%20high%20genetic%20heterogeneity.%20Genome-wide%20association%20studies%20have%20revealed%20a%20shared%20genetic%20background%20between%20POI%20and%20reproductive%20aging.%20NGS%20will%20provide%20a%20genetic%20diagnosis%20leading%20to%20genetic%5C%2Ftherapeutic%20counseling%3A%20first%2C%20defects%20in%20meiosis%20or%20DNA%20repair%20genes%20may%20predispose%20to%20tumors%3B%20and%20second%2C%20specific%20gene%20defects%20may%20predict%20the%20risk%20of%20rapid%20loss%20of%20a%20persistent%20ovarian%20reserve%2C%20an%20important%20determinant%20in%20fertility%20preservation.%20Indeed%2C%20a%20recent%20innovative%20treatment%20of%20POI%20by%20in%20vitro%20activation%20of%20dormant%20follicles%20proved%20to%20be%20successful.%22%2C%22date%22%3A%222018-06%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tem.2018.03.010%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221879-3061%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%22VPXIV2RV%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Penrad-Mobayed%20et%20al.%22%2C%22parsedDate%22%3A%222018-05-08%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPenrad-Mobayed%2C%20M.%2C%20Perrin%2C%20C.%2C%20L%26%23×2019%3BH%26%23xF4%3Bte%2C%20D.%2C%20Contremoulins%2C%20V.%2C%20Lepesant%2C%20J.-A.%2C%20Boizet-Bonhoure%2C%20B.%2C%20Poulat%2C%20F.%2C%20Baudin%2C%20X.%2C%20%26amp%3B%20Veitia%2C%20R.%20A.%20%282018%29.%20A%20role%20for%20SOX9%20in%20post-transcriptional%20processes%3A%20insights%20from%20the%20amphibian%20oocyte.%20%26lt%3Bi%26gt%3BScientific%20Reports%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B8%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%207191.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-018-25356-1%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-018-25356-1%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20role%20for%20SOX9%20in%20post-transcriptional%20processes%3A%20insights%20from%20the%20amphibian%20oocyte%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Penrad-Mobayed%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C.%22%2C%22lastName%22%3A%22Perrin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%22%2C%22lastName%22%3A%22L%27H%5Cu00f4te%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22V.%22%2C%22lastName%22%3A%22Contremoulins%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J.-A.%22%2C%22lastName%22%3A%22Lepesant%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B.%22%2C%22lastName%22%3A%22Boizet-Bonhoure%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22F.%22%2C%22lastName%22%3A%22Poulat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22X.%22%2C%22lastName%22%3A%22Baudin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20A.%22%2C%22lastName%22%3A%22Veitia%22%7D%5D%2C%22abstractNote%22%3A%22Sox9%20is%20a%20member%20of%20the%20gene%20family%20of%20SOX%20transcription%20factors%2C%20which%20is%20highly%20conserved%20among%20vertebrates.%20It%20is%20involved%20in%20different%20developmental%20processes%20including%20gonadogenesis.%20In%20all%20amniote%20species%20examined%20thus%20far%2C%20Sox9%20is%20expressed%20in%20the%20Sertoli%20cells%20of%20the%20male%20gonad%2C%20suggesting%20an%20evolutionarily%20conserved%20role%20in%20testis%20development.%20However%2C%20in%20the%20anamniotes%2C%20fishes%20and%20amphibians%2C%20it%20is%20also%20expressed%20in%20the%20oocyte%20but%20the%20significance%20of%20such%20an%20expression%20remains%20to%20be%20elucidated.%20Here%2C%20we%20have%20investigated%20the%20nuclear%20localization%20of%20the%20SOX9%20protein%20in%20the%20oocyte%20of%20three%20amphibian%20species%2C%20the%20urodelan%20Pleurodeles%20waltl%2C%20and%20two%20anurans%2C%20Xenopus%20laevis%20and%20Xenopus%20tropicalis.%20We%20demonstrate%20that%20SOX9%20is%20associated%20with%20ribonucleoprotein%20%28RNP%29%20transcripts%20of%20lampbrush%20chromosomes%20in%20an%20RNA-dependent%20manner.%20This%20association%20can%20be%20visualized%20by%20Super-resolution%20Structured%20Illumination%20Microscopy%20%28SIM%29.%20Our%20results%20suggest%20that%20SOX9%2C%20known%20to%20bind%20DNA%2C%20also%20carries%20an%20additional%20function%20in%20the%20posttranscriptional%20processes.%20We%20also%20discuss%20the%20significance%20of%20the%20acquisition%20or%20loss%20of%20Sox9%20expression%20in%20the%20oocyte%20during%20evolution%20at%20the%20transition%20between%20anamniotes%20and%20amniotes.%22%2C%22date%22%3A%222018-05-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41598-018-25356-1%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222045-2322%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%223ZUDK9KF%22%5D%2C%22dateModified%22%3A%222023-05-02T12%3A08%3A31Z%22%7D%7D%5D%7D
Veitia, R. A., Herman, L., Legois, B., Claret, S., Zider, A., & Todeschini, A.-L. (2025). Deciphering the impact of AKT1 pathogenic variants in Juvenile Granulosa Cell Tumors Using a Drosophila model.
Molecular & Cellular Proteomics,
0(0).
https://doi.org/10.1016/j.mcpro.2025.101466
Veitia, R. A., Cowles, J. D., & Caburet, S. (2025). Reclassifying NOBOX variants in primary ovarian insufficiency cases with a corrected gene model and a novel quantitative framework.
Human Reproduction, deaf058.
https://doi.org/10.1093/humrep/deaf058
Veitia, R. A. (2025). Rethinking transcription factor dynamics and transcription regulation in eukaryotes.
Trends in Biochemical Sciences.
https://doi.org/10.1016/j.tibs.2025.01.012
Veitia, R. A., Zschocke, J., & Birchler, J. A. (2025). Gene Dosage Sensitivity and Human Genetic Diseases.
Journal of Inherited Metabolic Disease,
48(4), e70058.
https://doi.org/10.1002/jimd.70058
Veitia, R. A. (2024). Emerging links between phase separation and transcription factor haploinsufficiency.
Trends in Genetics.
https://doi.org/10.1016/j.tig.2024.10.001
Herman, L., Amo, A., Legois, B., Di Carlo, C., Veitia, R. A., & Todeschini, A.-L. (2024). A cellular model provides insights into the pathogenicity of the oncogenic FOXL2 somatic variant p.Cys134Trp.
British Journal of Cancer, 1–10.
https://doi.org/10.1038/s41416-024-02613-x
Zutterling, C., Todeschini, A.-L., Fourmy, D., Busso, D., Veaute, X., Ducongé, F., & Veitia, R. A. (2023). The forkhead DNA-binding domain binds specific G2-rich RNA sequences.
Nucleic Acids Research, gkad994.
https://doi.org/10.1093/nar/gkad994
Pozzi, C., Vanet, A., Francesconi, V., Tagliazucchi, L., Tassone, G., Venturelli, A., Spyrakis, F., Mazzorana, M., Costi, M. P., & Tonelli, M. (2023). Antitarget, Anti-SARS-CoV-2 Leads, Drugs, and the Drug Discovery-Genetics Alliance Perspective.
Journal of Medicinal Chemistry,
66(6), 3664–3702.
https://doi.org/10.1021/acs.jmedchem.2c01229
Ariste, O., de la Grange, P., & Veitia, R. A. (2023). Recurrent missense variants in clonal hematopoiesis-related genes present in the general population.
Clinical Genetics,
103(2), 247–251.
https://doi.org/10.1111/cge.14259
Llano, E., Todeschini, A. L., Felipe-Medina, N., Corte-Torres, M. D., Condezo, Y. B., Sanchez-Martin, M., López-Tamargo, S., Astudillo, A., Puente, X. S., Pendas, A. M., & Veitia, R. A. (2023). The Oncogenic FOXL2 C134W Mutation Is a Key Driver of Granulosa Cell Tumors.
Cancer Research,
83(2), 239–250.
https://doi.org/10.1158/0008-5472.CAN-22-1880
Veitia, R. A. (2022). Who ever thought genetic mutations were random?
Trends in Plant Science,
27(8), 733–735.
https://doi.org/10.1016/j.tplants.2022.03.003
Franca, M. M., Condezo, Y. B., Elzaiat, M., Felipe-Medina, N., Sánchez-Sáez, F., Muñoz, S., Sainz-Urruela, R., Martín-Hervás, M. R., García-Valiente, R., Sánchez-Martín, M. A., Astudillo, A., Mendez, J., Llano, E., Veitia, R. A., Mendonca, B. B., & Pendás, A. M. (2022). A truncating variant of RAD51B associated with primary ovarian insufficiency provides insights into its meiotic and somatic functions.
Cell Death and Differentiation.
https://doi.org/10.1038/s41418-022-01021-z
Barakizou, H., Souha, G., Kamoun, T., Mehdi, M., Amary, F., Huma, Z., Todeschini, A., Veitia, R., & Donaldson, M. (2022). Precocious Pseudo-puberty in a Two-year-old Girl, Presenting with Bilateral Ovarian Enlargement and Progressing to Unilateral Juvenile Granulosa Cell Tumour.
Journal of Clinical Research in Pediatric Endocrinology,
14(1), 107–113.
https://doi.org/10.4274/jcrpe.galenos.2021.2021.0039
Veitia, R. A., & Innan, H. (2022). Pathogenic “germline” variants associated with myeloproliferative disorders in apparently normal individuals: Inherited or acquired genetic alterations?
Clinical Genetics,
101(3), 371–374.
https://doi.org/10.1111/cge.14104
Veitia, R. A., & Birchler, J. A. (2022). Gene-dosage issues: a recurrent theme in whole genome duplication events.
Trends in Genetics: TIG,
38(1), 1–3.
https://doi.org/10.1016/j.tig.2021.06.006
Pilsworth, J. A., Todeschini, A.-L., Neilson, S. J., Cochrane, D. R., Lai, D., Anttonen, M., Heikinheimo, M., Huntsman, D. G., & Veitia, R. A. (2021). FOXL2 in adult-type granulosa cell tumour of the ovary: oncogene or tumour suppressor gene?
The Journal of Pathology,
255(3), 225–231.
https://doi.org/10.1002/path.5771
Veitia, R. A. (2021). A reply to: Longitudinal changes in the frequency of mosaic chromosome Y loss in peripheral blood cells of aging men varies profoundly between individuals.
European Journal of Human Genetics: EJHG,
29(9), 1321–1322.
https://doi.org/10.1038/s41431-020-00801-w
Veitia, R. A., Pilsworth, J., Todeschini, A.-L., & Huntsman, D. (2021). Reply to “An alternative miRISC targets a cancer-associated coding sequence mutation in FOXL2.”
The EMBO Journal,
40(16), e107517.
https://doi.org/10.15252/embj.2020107517
Bermúdez-Guzmán, L., & Veitia, R. A. (2021). Insights into the pathogenicity of missense variants in the forkhead domain of FOX proteins underlying Mendelian disorders.
Human Genetics,
140(7), 999–1010.
https://doi.org/10.1007/s00439-021-02267-2
Herman, L., Todeschini, A.-L., & Veitia, R. A. (2021). Forkhead Transcription Factors in Health and Disease.
Trends in Genetics: TIG,
37(5), 460–475.
https://doi.org/10.1016/j.tig.2020.11.003
Innan, H., Vaiman, D., & Veitia, R. A. (2021). Predictable increase in female reproductive window: A simple model connecting age of reproduction, menopause, and longevity.
BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology,
43(5), e2000233.
https://doi.org/10.1002/bies.202000233
Herman, L., Legois, B., Todeschini, A.-L., & Veitia, R. A. (2021). Genomic exploration of the targets of FOXL2 and ESR2 unveils their implication in cell migration, invasion, and adhesion.
FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology,
35(4), e21355.
https://doi.org/10.1096/fj.202002444R
Caburet, S., Heddar, A., Dardillac, E., Creux, H., Lambert, M., Messiaen, S., Tourpin, S., Livera, G., Lopez, B. S., & Misrahi, M. (2021). Homozygous hypomorphic BRCA2 variant in primary ovarian insufficiency without cancer or Fanconi anaemia trait.
Journal of Medical Genetics,
58(2), 125–134.
https://doi.org/10.1136/jmedgenet-2019-106672
Veitia, R. A. (2021). Clinical Genetics paving the way to the future.
Clinical Genetics,
99(2), 217–218.
https://doi.org/10.1111/cge.13899
Birchler, J. A., & Veitia, R. A. (2021). One Hundred Years of Gene Balance: How Stoichiometric Issues Affect Gene Expression, Genome Evolution, and Quantitative Traits.
Cytogenetic and Genome Research,
161(10–11), 529–550.
https://doi.org/10.1159/000519592
Picolo, F., Grandchamp, A., Piégu, B., Rolland, A. D., Veitia, R. A., & Monget, P. (2021). Genes Encoding Teleost Orthologs of Human Haploinsufficient and Monoallelically Expressed Genes Remain in Duplicate More Frequently Than the Whole Genome.
International Journal of Genomics,
2021, 9028667.
https://doi.org/10.1155/2021/9028667
Elzaiat, M., Flatters, D., Sierra-Díaz, D. C., Legois, B., Laissue, P., & Veitia, R. A. (2020). DHH pathogenic variants involved in 46,XY disorders of sex development differentially impact protein self-cleavage and structural conformation.
Human Genetics,
139(11), 1455–1470.
https://doi.org/10.1007/s00439-020-02189-5
Alvarez-Mora, M. I., Todeschini, A.-L., Caburet, S., Perets, L. P., Mila, M., Younis, J. S., Shalev, S., & Veitia, R. A. (2020). An exome-wide exploration of cases of primary ovarian insufficiency uncovers novel sequence variants and candidate genes.
Clinical Genetics,
98(3), 293–298.
https://doi.org/10.1111/cge.13803
Felipe-Medina, N., Caburet, S., Sánchez-Sáez, F., Condezo, Y. B., de Rooij, D. G., Gómez-H, L., Garcia-Valiente, R., Todeschini, A. L., Duque, P., Sánchez-Martin, M. A., Shalev, S. A., Llano, E., Veitia, R. A., & Pendás, A. M. (2020). A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1.
eLife,
9, e56996.
https://doi.org/10.7554/eLife.56996
Govindaraju, D. R., Innan, H., & Veitia, R. A. (2020). The Muller’s Ratchet and Aging.
Trends in Genetics: TIG,
36(6), 395–402.
https://doi.org/10.1016/j.tig.2020.02.004
Veitia, R. A. (2020). Primary ovarian insufficiency, meiosis and DNA repair.
Biomedical Journal,
43(2), 115–123.
https://doi.org/10.1016/j.bj.2020.03.005
Innan, H., Veitia, R., & Govindaraju, D. R. (2020). Genetic and epigenetic Muller’s ratchet as a mechanism of frailty and morbidity during aging: a demographic genetic model.
Human Genetics,
139(3), 409–420.
https://doi.org/10.1007/s00439-019-02067-9
Benayoun, B. A., & Veitia, R. A. (2020). Special issue on “Molecular genetics of aging and longevity”: a critical time in the field of geroscience.
Human Genetics,
139(3), 275–276.
https://doi.org/10.1007/s00439-020-02125-7
Grassmann, F., International AMD Genomics Consortium (IAMDGC), Weber, B. H. F., & Veitia, R. A. (2020). Insights into the loss of the Y chromosome with age in control individuals and in patients with age-related macular degeneration using genotyping microarray data.
Human Genetics,
139(3), 401–407.
https://doi.org/10.1007/s00439-019-02029-1
Penrad-Mobayed, M., Perrin, C., Herman, L., Todeschini, A.-L., Nigon, F., Cosson, B., Caburet, S., & Veitia, R. A. (2020). Conventional and unconventional interactions of the transcription factor FOXL2 uncovered by a proteome-wide analysis.
FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology,
34(1), 571–587.
https://doi.org/10.1096/fj.201901573R
Shi, X., Chen, C., Yang, H., Hou, J., Ji, T., Cheng, J., Veitia, R. A., & Birchler, J. A. (2020). The Gene Balance Hypothesis: Epigenetics and Dosage Effects in Plants.
Methods in Molecular Biology (Clifton, N.J.),
2093, 161–171.
https://doi.org/10.1007/978-1-0716-0179-2_12
Veitia, R. A. (2019). MIRAGE Syndrome: Phenotypic Rescue by Somatic Mutation and Selection.
Trends in Molecular Medicine,
25(11), 937–940.
https://doi.org/10.1016/j.molmed.2019.08.008
Johnson, A. F., Nguyen, H. T., & Veitia, R. A. (2019). Causes and effects of haploinsufficiency.
Biological Reviews of the Cambridge Philosophical Society,
94(5), 1774–1785.
https://doi.org/10.1111/brv.12527
Veitia, R. A. (2019). AFF3: a new player in maintaining XIST monoallelic expression.
Journal of Molecular Cell Biology,
11(9), 723–724.
https://doi.org/10.1093/jmcb/mjy082
Veitia, R. A. (2019). Darwinian selection within an individual or somatic selection: facts and models.
Journal of Molecular Cell Biology,
11(8), 719–722.
https://doi.org/10.1093/jmcb/mjz014
Veitia, R. A. (2019). DNA Content, Cell Size, and Cell Senescence.
Trends in Biochemical Sciences,
44(8), 645–647.
https://doi.org/10.1016/j.tibs.2019.04.013
Elzaiat, M., Herman, L., Legois, B., Léger, T., Todeschini, A.-L., & Veitia, R. A. (2019). High-throughput Exploration of the Network Dependent on AKT1 in Mouse Ovarian Granulosa Cells.
Molecular & Cellular Proteomics: MCP,
18(7), 1307–1319.
https://doi.org/10.1074/mcp.RA119.0014613
Veitia, R. A. (2019). Further quantitative insights into the decrease of heteroplasmy of m.3243A>G with age in leukocytes.
Clinical Genetics,
95(4), 542–543.
https://doi.org/10.1111/cge.13496
Caburet, S., Todeschini, A.-L., Petrillo, C., Martini, E., Farran, N. D., Legois, B., Livera, G., Younis, J. S., Shalev, S., & Veitia, R. A. (2019). A truncating MEIOB mutation responsible for familial primary ovarian insufficiency abolishes its interaction with its partner SPATA22 and their recruitment to DNA double-strand breaks.
EBioMedicine,
42, 524–531.
https://doi.org/10.1016/j.ebiom.2019.03.075
Ouimette, J.-F., Rougeulle, C., & Veitia, R. A. (2019). Three-dimensional genome architecture in health and disease.
Clinical Genetics,
95(2), 189–198.
https://doi.org/10.1111/cge.13219
Birchler, J. A., & Veitia, R. A. (2019). Genomic Balance and Speciation.
Epigenetics Insights,
12, 2516865719840291.
https://doi.org/10.1177/2516865719840291
Veitia, R. A. (2018). On the loss of human sex chromosomes in lymphocytes with age: a quantitative treatment.
European Journal of Human Genetics: EJHG,
26(12), 1875–1878.
https://doi.org/10.1038/s41431-018-0225-0
Veitia, R. A. (2018). Dosage effects in morphogenetic gradients of transcription factors: insights from a simple mathematical model. Journal of Genetics, 97(2), 365–370.
Huhtaniemi, I., Hovatta, O., La Marca, A., Livera, G., Monniaux, D., Persani, L., Heddar, A., Jarzabek, K., Laisk-Podar, T., Salumets, A., Tapanainen, J. S., Veitia, R. A., Visser, J. A., Wieacker, P., Wolczynski, S., & Misrahi, M. (2018). Advances in the Molecular Pathophysiology, Genetics, and Treatment of Primary Ovarian Insufficiency.
Trends in Endocrinology and Metabolism: TEM,
29(6), 400–419.
https://doi.org/10.1016/j.tem.2018.03.010
Penrad-Mobayed, M., Perrin, C., L’Hôte, D., Contremoulins, V., Lepesant, J.-A., Boizet-Bonhoure, B., Poulat, F., Baudin, X., & Veitia, R. A. (2018). A role for SOX9 in post-transcriptional processes: insights from the amphibian oocyte.
Scientific Reports,
8(1), 7191.
https://doi.org/10.1038/s41598-018-25356-1
Theses
2016-2020 : Laëtitia Herman, doctorat soutenu le 22/09/2020 « Caractérisation moléculaire du rôle de FOXL2 et de ses partenaires dans l’ovaire sain et pathologique »
2012-2015 : Laurianne Bessière, doctorat soutenu le 16/09/2015 « Exploration génomique et fonctionnelle des tumeurs des cellules de la granulosa ovarienne »
2009-2013 : Adrien Georges, doctorat soutenu le 28/06/2013 « FOXL2 : un déterminant de la signalisation dans l’ovaire »
2007-2011 : Bérénice Benayoun, doctorat soutenu le 28/01/2011 « FOXL2 : un facteur de transcription essentiel de l’ovaire, à l’interface entre la réponse au stress cellulaire et la suppression tumorale »
2006-2009 : Aurélie Dipiétromaria, doctorat soutenu le 28/09/2009 « Etudes des conséquences fonctionnelles des mutations de FOXL2 : gène impliqué dans le syndrome du BPES »
2005-2008 : Franck Batista Pelaez, doctorat soutenu le 13/06/2008 « Les cibles transcriptionnelles de FOXL2, un facteur de transcription impliqué dans le développement et le maintien de la fonction ovarienne »
2005-2008 : Lara Moumné, doctorat soutenu le 14/05/2008 « Etude fonctionnelle des mutations affectant FOXL2 : un acteur clé du développement de l’ovaire chez les vertébrés »
2002-2006 : Julie Cocquet doctorat soutenu le //2006 « Evolution, expression et structure de FOXL2, un gène impliqué dans la fertilité féminine »
Collaborations
Etudes sur les tumeurs des cellules de la granulosa ovarienne adultes (AGCTs) et sur le facteur de transcription FOXL2 : Equipe d’E. de Baere, Ghent Belgique. Équipe de N. Binart, Kremlin-Bicêtre, France. F. Poulat, Montpellier, France. Equipe de M. Anttonen, Helsinki, Finlande. Equipe D. Huntsman, Vancouver, Canada. Equipe d’Alberto Pendas, Salamanca, Espagne.
Etudes sur les tumeurs des cellules de la granulosa ovarienne juvéniles (JGCTs) et sur l’oncoprotéine AKT1 : S. Sarnacki et L. Galmiche, Paris. N. Kalfa et C. Sultan, Montpellier. J. Prat et E. d’Angelo, Barcelona, Espagne. Malcolm Donaldson, Glasgow, United Kingdom.
Etudes sur les gènes impliqués dans les insuffisances ovariennes prématurées : Maria Isabel Alvarez-Mora et Montserrat Mila, Barcelona, Espagne. Equipe de Gabriel Livera, Fontenay aux Roses, France. Johnny S. Younis, Safed, Israel. Equipe de Stavit Shalev, Afula, Israel. Equipe d’Alberto Pendas, Salamanca, Espagne
Grants - funding
1) ANR (National Agency for Research) : OVARYPROTECT : Interaction entre la signalisation PKA et TRIM28 dans la maintenance et la pathologie ovarienne
2) GEFLUC (Groupement des Entreprises Françaises dans la Lutte contre le Cancer): Ovarian cancer: FOXL2-C134W impact on cell-cell or cell-ECM adhesion.
News
Job