Chromosomal Domains and DNA Replication
MARIE-NOËLLE PRIOLEAU
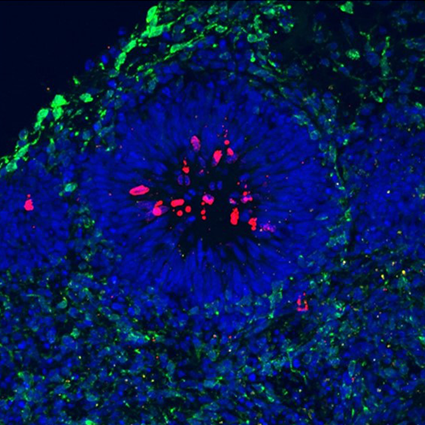
During development, cells actively divide and gradually organize chromosomal expression domains. Before each division, they must copy their genome with a high degree of accuracy. An error in this process can lead to cell death or genetic instability that can lead to the development of cancer cells. This crucial step is called DNA replication. It begins at many specific sites in the genome—about 100,000 in humans—called replication origins.
Keywords: DNA replication, replication initiation, G-quadruplex, chromatin, fragile sites, cortical organoid
+33 (0)1 57 27 81 02 Contact @MariePrioleau
Initiation of replication in vertebrates
In eukaryotes, the activation of replication origins is finely regulated in space and time during the S phase of the cell cycle. Our team seeks to identify the molecular mechanisms that establish and control this program in vertebrates. We are also interested in how two fundamental processes, transcription and replication, can occur simultaneously on the same DNA molecule in a coordinated manner. Indeed, conflicts between these two machineries can cause genetic instability.
In order to obtain a comprehensive view of human genome replication, we are developing high-throughput approaches to map replication origins, their activation timing, and regions of the genome prone to damage (Figure 1). The results obtained from these genomic analyses are then validated using genetic approaches.
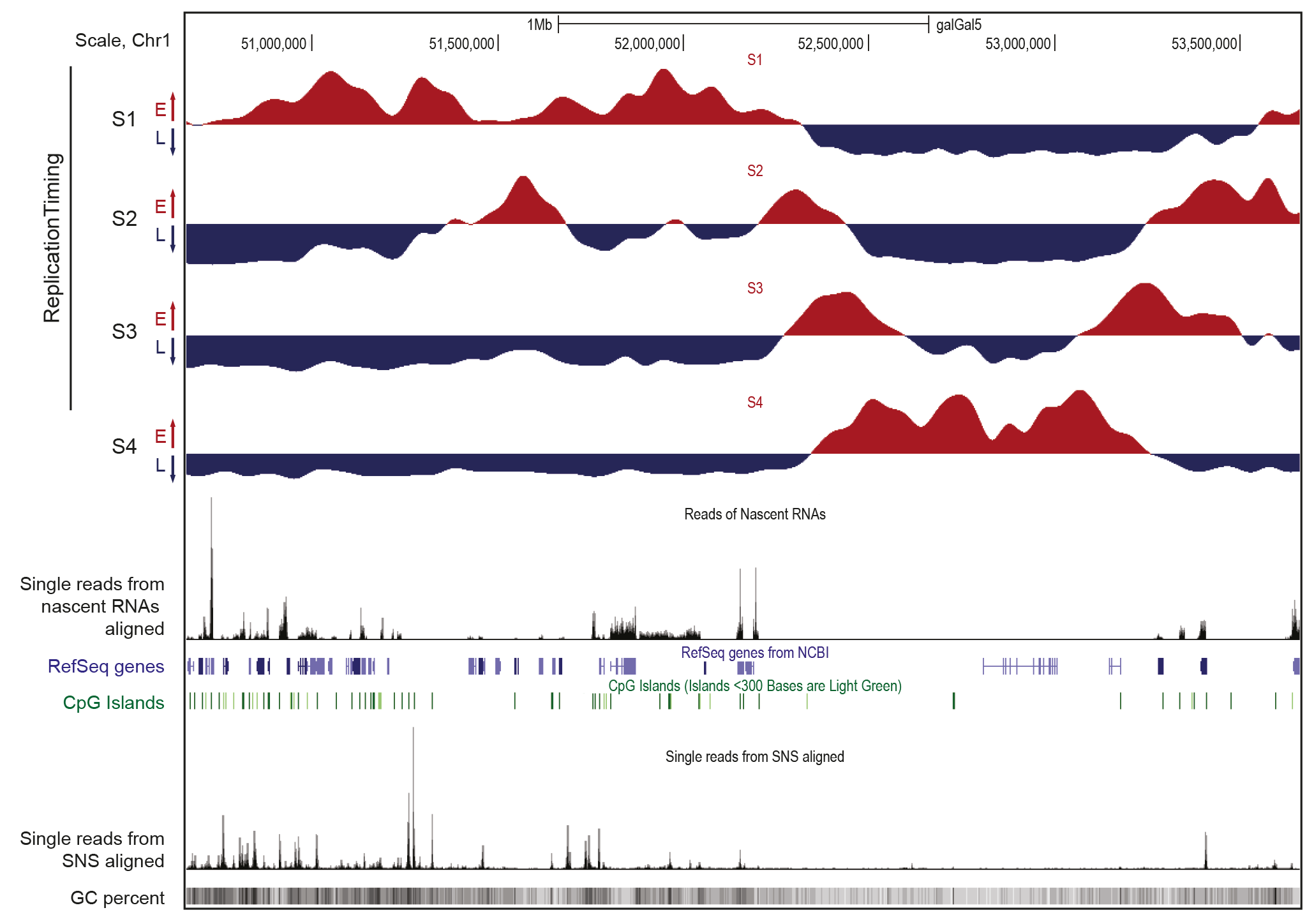 Figure 1. Representative features of early and late replication domains
Figure 1. Representative features of early and late replication domains
For many years, we have been using an avian cell model, the DT40 lymphoid cell line, which is characterized by particularly efficient homologous recombination. This model has enabled us to demonstrate that DNA structures called G-quadruplexes play a key role in the initiation of replication. We are now implementing the use of a human lymphoid cell line, TK6, which we are genetically modifying using the CRISPR/Cas9 tool. In addition, in order to study the impact of replication dysfunction on tissue development, we are developing analyses on human cortical organoids.
Our current projects aim to understand how G-quadruplexes facilitate the recruitment of the replication machinery, as well as the reciprocal interactions between replication origins and transcription.
Members
 Theo BARET, PhD student, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Theo BARET, PhD student, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Caroline DONCARLI, Biology engineer, PRIOLEAU LAB+33 (0)1 57 27 81 24, room 520B
Caroline DONCARLI, Biology engineer, PRIOLEAU LAB+33 (0)1 57 27 81 24, room 520B William MALLETT, Biology engineer, PRIOLEAU LAB
William MALLETT, Biology engineer, PRIOLEAU LAB Kathrin MARHEINEKE, Researcher, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Kathrin MARHEINEKE, Researcher, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Aurélie MASSON, PhD student, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Aurélie MASSON, PhD student, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
To contact a member of the team by e-mail: name.surname@ijm.fr
1) Transcription Represses Origin Activity in a Late-Replicating Fragile Site. Mandelbrojt, J., Tonnerre-Doncarli, C., Masson, A., Baret, T., Debatisse, M., and Prioleau, M.-N. (2025). Preprint at bioRxiv, https://doi.org/10.1101/2025.08.01.668051.
2) Dual DNA replication modes: varying fork speeds and initiation rates within the spatial replication program in Xenopus. Ciardo D, Haccard O, de Carli F, Hyrien O, Goldar A, Marheineke K. Nucleic Acids Res. 2025 Jan 24;53(3):gkaf007. doi: 10.1093/nar/gkaf007.
3) Dimeric G-quadruplex motifs-induced NFRs determine strong replication origins in vertebrates.Poulet-Benedetti J, Tonnerre-Doncarli C, Valton AL, Laurent M, Gérard M, Barinova N, Parisis N, Massip F, Picard F, Prioleau MN. Nat Commun. 2023 Aug 10;14(1):4843. doi: 10.1038/s41467-023-40441-4.
4) Polo-like kinase 1 (Plk1) regulates DNA replication origin firing and interacts with Rif1 in Xenopus. Ciardo D, Haccard O, Narassimprakash H, Cornu D, Guerrera IC, Goldar A, Marheineke K. Nucleic Acids Res. 2021 Sep 27;49(17):9851-9869. doi: 10.1093/nar/gkab756.
5) Clustering of strong replicators associated with active promoters is sufficient to establish an early-replicating domain. Brossas C, Valton AL, Venev SV, Chilaka S, Counillon A, Laurent M, Goncalves C, Duriez B, Picard F, Dekker J, Prioleau MN. EMBO J. 2020 Nov 2;39(21):e99520. doi: 10.15252/embj.201899520. Epub 2020 Sep 16. PMID: 32935369
6) Replication dynamics of individual loci in single living cells reveal changes in the degree of replication stochasticity through S phase. Duriez B, Chilaka S, Bercher JF, Hercul E, Prioleau MN. Nucleic Acids Res. 2019 Jun 4;47(10):5155-5169. doi: 10.1093/nar/gkz220. PMID: 30926993
7) Evolution of replication origins in vertebrate genomes: rapid turnover despite selective constraints. Massip F, Laurent M, Brossas C, Fernández-Justel JM, Gómez M, Prioleau MN, Duret L, Picard F. Nucleic Acids Res. 2019 Jun 4;47(10):5114-5125. doi: 10.1093/nar/gkz182. PMID: 30916335
8) Transcription-dependent regulation of replication dynamics modulates genome stability. Blin M, Le Tallec B, Nähse V, Schmidt M, Brossas C, Millot GA, Prioleau MN, Debatisse M. Nat Struct Mol Biol. 2019 Jan;26(1):58-66. doi: 10.1038/s41594-018-0170-1. Epub 2018 Dec 31. PMID: 30598553
9) The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. Picard F, Cadoret JC, Audit B, Arneodo A, Alberti A, Battail C, Duret L, Prioleau MN. PLoS Genet. 2014 May 1;10(5):e1004282. doi: 10.1371/journal.pgen.1004282. eCollection 2014 May. PMID: 24785686
10) G4 motifs affect origin positioning and efficiency in two vertebrate replicators. Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, Riou JF, Prioleau MN. EMBO J. 2014 Apr 1;33(7):732-46. doi: 10.1002/embj.201387506. Epub 2014 Feb 12. PMID: 24521668
Publications
- « Rôle de l’insulateur 5’HS4 du poulet dans la régulation de la réplication » soutenue en Juin 2009 par Vahideh Hassan-Zadeh.
- « Mécanismes moléculaires de l’initiation de la réplication » soutenue en Décembre 2010 par Françoise Meisch.
- « Identification de séquences cis-nucléiques nécessaires à l’initiation de la réplication chez les vertébrés » soutenue en Juin 2014 par Anne-Laure Valton.
- « Construction d’un domaine synthétique de réplication précoce et impact sur la structure chromatinienne et la permissivité transcriptionnelle » soutenue en Septembre 2015 par Caroline Tonnerre-Doncarli Brossas.
- « Mécanismes moléculaires impliqués dans la régulation du moment de déclenchement des origines de réplication » soutenue en Novembre 2016 par Antonin Counillon.
- « Rôle des G-quadruplexes dans la spécification des origines de réplication chez les vertébrés » soutenue en Septembre 2016 par Marc Laurent.
- « Etude moléculaire des éléments cis-régulateurs et de l’organisation de la chromatine des origines de réplication » soutenue en Septembre 2020 par Jérémy Poulet-Benedetti.
« Comment la transcription et le contexte chromosomique interfère avec une origine de réplication efficace ? » soutenue en décembre 2024 par Juliette Mandelbrojt
Nationals:
- Imagerie cellulaire et cytométrie en flux : Plateforme Imago-Seine, Institut Jacques Monod
- Organoïdes corticaux : Plateforme enSCORE, Labex Who am I ?
- Bio-informatique et bio-statistique : LBBE, UCB Lyon 1 (Laurent Duret) et ENS de Lyon (Franck Picard)
- G-quadruplex : Inserm U565, CNRS UMR 7196, MNHN, Paris, France (Patrizia Alberti, Carole Saintomé et Jean-François Riou)
- Sites Fragiles Communs : : Institut Gustave Roussy, Villejuif (Michelle Debatisse et Stéphane Koundrioukoff)
Internationals:
- Organisation tri-dimensionnelle du noyau : University of Massachusetts Medical School, Worcester, Etats-Unis (Anne-Laure Valton et Job Dekker)
- Rôle des G4s dans le positionnement des nucléosomes : Fox Chase Cancer center, Philadelphie, Etats-Unis et Moscow State University, Moscow, Russie (Vasily Studitsky).


