Cycle cellulaire et développement
LIONEL PINTARD
The Cell Cycle and Development team aims to acquire new knowledge to decipher the mechanisms of cell division to understand the mechanisms of cancer, a disease resulting from uncontrolled cell division.
The team mainly uses the nematode C. elegans as a model system and employs a multidisciplinary approach combining various approaches (biochemistry, genetics, imaging, proteomics) to ask questions at different scales, from the molecule to the organism. As the mechanisms of regulation of cell division are conserved between species, the team is also studying the emerging paradigms in C. elegans in human cells.
Keywords: Cell division – Mitosis – Meiosis –Kinases – Microtubule-severing enzymes – Nuclear envelope breakdown
+33 (0)1 57 27 80 89 Contact @ccdlab.bsky.social https://sites.google.com/site/pintardlab/
Our vision
Acquiring new knowledge to decipher how cells divide, which is key to better understand the mechanisms of cancer, a disease caused by uncontrolled cell division.
Context
Humans are roughly built from 1013 cells corresponding to 200 different cell types. All these cells are generated through cell divisions, starting from a single cell, the fertilized egg. To generate these large number of cells and maintain tissue homeostasis, the human body experiences as many as 1016 cell divisions in a lifetime. During each cell division, the genome must be faithfully replicated and equally segregated between the daughter cells during mitosis. Defects in these processes can have drastic consequences, leading to outcomes such as cell death, genome instability or deregulated growth, typical of cancer. Despite considerable progress, the mechanisms regulating cell division are incompletely understood. This lack of knowledge has considerably limited the development of innovative therapeutic approaches.
Research programm
I- Mechanisms regulating mitotic entry in space and time
Commitment to mitosis must be tightly coordinated with DNA replication to preserve genome integrity. Consistently, unscheduled mitosis may contribute to genetic instability. Entry into mitosis is controlled by evolutionarily conserved serine/threonine kinases (Aurora A, Polo-like kinase, Plk1) as well as counteracting phosphatases (PPases). How these kinase activities are regulated in space and time, and how they work in concert to trigger mitosis at the right time remain ill defined. In this context, we are investigating the
- Activation mechanism of key mitotic kinases (Aurora A, Polo-like kinase 1)
- Role of mitotic kinases in nuclear envelope breakdown (NEBD)
- Role and regulation of Mitotic kinases during asynchronous cell division
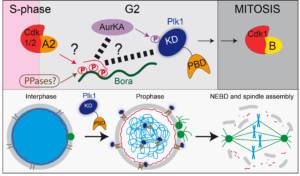
Figure 1: Bora-Aurora A (Aurka)-Plk1 axis during mitotic entry
II- Role and regulation of the microtubule-severing enzyme Katanin
Microtubules (MTs) are dynamic cytoskeleton polymers, which play a central role in cell division, morphogenesis, motility and signaling. Most MT regulatory proteins interact with the plus or minus end of microtubules and thereby control their polymerization and depolymerization rates. However, one class of MT regulator interacts with the MT lattice and severs MTs throughout their length, thereby controlling their size and density in the cell. Three evolutionarily conserved MT-severing enzymes have been identified: Fidgetin, Spastin and Katanin. Mutations of these enzymes have been linked to various defects and pathologies including developmental defects and neurodegenerative disorders. However, little is known about the precise mechanisms by which these enzymes sever MTs. Likewise, how these enzymes are regulated in space and time is poorly understood. We are currently focusing on deciphering the mode of action and regulation of Katanin, which is essential for female meiotic spindle assembly in C. elegans. We are focusing our work on the
- role of MT-severing in meiotic spindle assembly
- Mechanism of MT-severing by Katanin
- Regulation of Katanin-mediated MT-severing in space and time during development
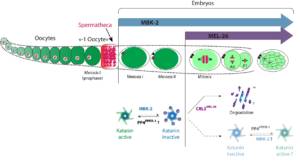 Figure 2: Role and regulation of Katanin during C. elegans development (From Joly et al. JCB 2020).
Figure 2: Role and regulation of Katanin during C. elegans development (From Joly et al. JCB 2020).
III- Cullin-RING E3-Ligases in cell division
Cullin-RING E3-ligases (CRL) represent the largest family of E3 ubiquitin-ligases targeting the degradation of key cell cycle regulators in space and time, contributing to the orderly progression of the cell division cycle. We are interested in understanding how these enzymes regulate cell cycle progression in a developmental context.
- CRL in the regulation of the Bora-Aurora-Plk1 pathway
- CRL in the regulation of Katanin activity
- CRL in the maintenance of DNA replication integrity
Methods
We use a multidisciplinary approach including biochemistry (reconstitution of enzymatic activities from purified components to dissect molecular mechanisms), genetics, live cell imaging, proteomics approaches using both human cells and the nematode C. elegans. Mechanisms regulating cell division are conserved between species such that emerging paradigm from C. elegans can be immediately investigated in human cells. Furthermore, C. elegans offers a number of practical advantages to study conserved pathways regulating cell division (Pintard & Bowerman, Wormbook, Genetics 2019).
Members









To contact a member of the team by e-mail: name.surname@ijm.fr

June, 2021 (c) Pintard Lab
From left to right: Anais, Griselda, Batool, Lucie, NicoT, Sylvia, Lionel, Eva, Lola, Anaelle, Emma, NicoJ
Roumbo, L., Ossareh-Nazari, B., Vigneron, S., Stefani, I., Van Hove, L., Legros, V., Chevreux, G., Lacroix, B., Castro, A., Joly, N., Lorca, T., & Pintard, L. (2025). The MAST kinase KIN-4 carries out mitotic entry functions of Greatwall in C. elegans. The EMBO Journal, 1–32. https://doi.org/10.1038/s44318-025-00364-w
Beaumale, E., Van Hove, L., Pintard, L., & Joly, N. (2024). Microtubule-binding domains in Katanin p80 subunit are essential for severing activity in C. elegans. Journal of Cell Biology, 223(4), e202308023. https://doi.org/10.1083/jcb.202308023
Nkombo Nkoula, S., Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Ayuso, C., Legros, V., Chevreux, G., Thomas, L., Seydoux, G., Askjaer, P., & Pintard, L. (2023). Mechanisms of nuclear pore complex disassembly by the mitotic Polo-like kinase 1 (PLK-1) in C. elegans embryos. Science Advances, 9(29), eadf7826. https://doi.org/10.1126/sciadv.adf7826
Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Joly, N., & Pintard, L. (2022). Cortical microtubule pulling forces contribute to the union of the parental genomes in the Caenorhabditis elegans zygote. ELife, 11, e75382. https://doi.org/10.7554/eLife.75382
Tavernier, N., Thomas, Y., Vigneron, S., Maisonneuve, P., Orlicky, S., Mader, P., Regmi, S. G., Van Hove, L., Levinson, N. M., Gasmi-Seabrook, G., Joly, N., Poteau, M., Velez-Aguilera, G., Gavet, O., Castro, A., Dasso, M., Lorca, T., Sicheri, F., & Pintard, L. (2021). Bora phosphorylation substitutes in trans for T-loop phosphorylation in Aurora A to promote mitotic entry. Nature Communications, 12(1), 1899. https://doi.org/10.1038/s41467-021-21922-w
Publications
2913254
GWSKYQWE
1
apa
50
date
desc
8954
https://www.ijm.fr/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22VJ8Z49XW%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Pillan%20et%20al.%22%2C%22parsedDate%22%3A%222026-01-28%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPillan%2C%20A.%2C%20Ormancey%2C%20P.%2C%20Ben%20Choug%2C%20C.%2C%20Orlicky%2C%20S.%2C%20Tavernier%2C%20N.%2C%20Van%20Hove%2C%20L.%2C%20Ossareh-Nazari%2C%20B.%2C%20Joly%2C%20N.%2C%20Sicheri%2C%20F.%2C%20Lorca%2C%20T.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282026%29.%20Molecular%20basis%20for%20the%20activation%20of%20Aurora%20A%20and%20Plk1%20kinases%20during%20mitotic%20entry.%20%26lt%3Bi%26gt%3BThe%20EMBO%20Journal%26lt%3B%5C%2Fi%26gt%3B.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00679-8%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00679-8%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Molecular%20basis%20for%20the%20activation%20of%20Aurora%20A%20and%20Plk1%20kinases%20during%20mitotic%20entry%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ana%5Cu00efs%22%2C%22lastName%22%3A%22Pillan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Philippine%22%2C%22lastName%22%3A%22Ormancey%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Celia%22%2C%22lastName%22%3A%22Ben%20Choug%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stephen%22%2C%22lastName%22%3A%22Orlicky%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Frank%22%2C%22lastName%22%3A%22Sicheri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thierry%22%2C%22lastName%22%3A%22Lorca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22The%20evolutionarily%20conserved%2C%20intrinsically%20disordered%20protein%20Bora%20is%20critical%20for%20initiating%20the%20activation%20of%20mitotic%20kinases.%20Once%20phosphorylated%20at%20Ser112%20by%20Cyclin%20A-Cdk1%20kinase%2C%20phospho-Bora%20activates%20unphosphorylated%20Aurora%20A%20kinase%20%28AURKA%29%2C%20directing%20it%20towards%20Polo-like%20kinase%201%20%28Plk1%29%2C%20thus%20promoting%20Cyclin%20B-Cdk1%20activation%20and%20mitotic%20entry.%20Here%2C%20by%20combining%20structural%20modeling%20and%20in%20vitro%20assays%2C%20we%20provide%20evidence%20that%20Bora%20wraps%20around%20the%20N-terminal%20lobe%20of%20AURKA%20to%20position%20its%20phospho-Ser112%20near%20AURKA%5Cu2019s%20T-loop%2C%20mimicking%20T-loop%20phosphorylation.%20Additionally%2C%20Bora%20transiently%20interacts%20with%20the%20%5Cu03b1C%20helix%20of%20the%20Plk1%20kinase%20domain%20through%20a%20conserved%20motif%2C%20guiding%20AURKA%20activity%20towards%20the%20Plk1%20T-loop%2C%20which%20is%20otherwise%20impervious%20to%20phosphorylation%20by%20AURKA.%20We%20highlight%20the%20importance%20of%20this%20motif%20for%20Bora%20function%20in%20vitro%20and%20during%20mitotic%20entry%20in%20Xenopus%20laevis%20egg%20extracts.%20Our%20results%20reveal%20critical%20molecular%20details%20of%20mitotic%20kinase%20activation%2C%20which%20could%20lead%20to%20the%20development%20of%20pathway-specific%20inhibitors.%22%2C%22date%22%3A%222026-01-28%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs44318-025-00679-8%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00679-8%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221460-2075%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222026-01-30T09%3A10%3A44Z%22%7D%7D%2C%7B%22key%22%3A%22YNT9FYUT%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Joly%20and%20Pintard%22%2C%22parsedDate%22%3A%222025-11-19%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BJoly%2C%20N.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282025%29.%20Intramolecular%20regulation%20of%20the%20MT-severing%20enzyme%20Katanin%20prevents%20futile%20ATP%20hydrolysis.%20%26lt%3Bi%26gt%3BJournal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B225%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20e202506192.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202506192%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202506192%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Intramolecular%20regulation%20of%20the%20MT-severing%20enzyme%20Katanin%20prevents%20futile%20ATP%20hydrolysis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Microtubule-severing%20enzymes%20are%20evolutionarily%20conserved%20AAA-ATPases%20that%20sever%20microtubules%2C%20thereby%20regulating%20diverse%20microtubule-dependent%20cellular%20processes.%20How%20these%20enzymes%20couple%20Microtubule%20binding%20with%20ATP%20hydrolysis%20to%20trigger%20microtubule-remodeling%20remains%20poorly%20understood.%20Using%20Caenorhabditiselegans%20Katanin%2C%20which%20contains%20the%20MEI-1%20catalytic%20AAA%2B%20p60%20and%20MEI-2%20p80-like%20regulatory%20subunits%2C%20we%20identify%20a%20critical%20regulatory%20role%20of%20the%20N-terminal%20domain%20of%20MEI-1%20in%20Katanin%20regulation.%20We%20demonstrate%20this%20domain%20represses%20the%20AAA%2B%20core%20in%20cis%2C%20limiting%20ATP%20hydrolysis%20and%20preventing%20interaction%20with%20tubulin%20C-terminal%20tails%20in%20the%20absence%20of%20MEI-2.%20Strikingly%2C%20MEI-1%20lacking%20its%20N%20terminus%20is%20constitutively%20active%2C%20enabling%20identification%20of%20pore%20residues%20critical%20for%20sensing%20microtubule%20C-terminal%20tails%20and%20relaying%20this%20signal%20to%20the%20AAA%2B%20core.%20These%20findings%20reveal%20how%20Katanin%20activation%20is%20coupled%20to%20microtubule%20binding%2C%20thereby%20avoiding%20futile%20ATP%20hydrolysis.%20Given%20Katanin%5Cu2019s%20evolutionary%20conservation%2C%20our%20work%20provides%20a%20mechanistic%20framework%20for%20its%20regulation%20in%20other%20organisms%2C%20with%20broader%20implications%20for%20human%20pathologies%2C%20including%20neurodegeneration%20and%20cancer.%22%2C%22date%22%3A%222025-11-19%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202506192%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202506192%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220021-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222025-11-25T15%3A20%3A51Z%22%7D%7D%2C%7B%22key%22%3A%224NIR3VJ9%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Roumbo%20et%20al.%22%2C%22parsedDate%22%3A%222025-02-17%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BRoumbo%2C%20L.%2C%20Ossareh-Nazari%2C%20B.%2C%20Vigneron%2C%20S.%2C%20Stefani%2C%20I.%2C%20Van%20Hove%2C%20L.%2C%20Legros%2C%20V.%2C%20Chevreux%2C%20G.%2C%20Lacroix%2C%20B.%2C%20Castro%2C%20A.%2C%20Joly%2C%20N.%2C%20Lorca%2C%20T.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282025%29.%20The%20MAST%20kinase%20KIN-4%20carries%20out%20mitotic%20entry%20functions%20of%20Greatwall%20in%20C.%20elegans.%20%26lt%3Bi%26gt%3BThe%20EMBO%20Journal%26lt%3B%5C%2Fi%26gt%3B%2C%201%26%23×2013%3B32.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00364-w%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00364-w%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20MAST%20kinase%20KIN-4%20carries%20out%20mitotic%20entry%20functions%20of%20Greatwall%20in%20C.%20elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ludivine%22%2C%22lastName%22%3A%22Roumbo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Suzanne%22%2C%22lastName%22%3A%22Vigneron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ioanna%22%2C%22lastName%22%3A%22Stefani%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22V%5Cu00e9ronique%22%2C%22lastName%22%3A%22Legros%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Guillaume%22%2C%22lastName%22%3A%22Chevreux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Benjamin%22%2C%22lastName%22%3A%22Lacroix%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thierry%22%2C%22lastName%22%3A%22Lorca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22MAST-like%2C%20or%20Greatwall%20%28Gwl%29%2C%20an%20atypical%20protein%20kinase%20related%20to%20the%20evolutionarily%20conserved%20MAST%20kinase%20family%2C%20is%20crucial%20for%20cell%20cycle%20control%20during%20mitotic%20entry.%20Mechanistically%2C%20Greatwall%20is%20activated%20by%20Cyclin%20B-Cdk1%20phosphorylation%20of%20a%20550%20amino%20acids-long%20insertion%20in%20its%20atypical%20activation%20segment.%20Subsequently%2C%20Gwl%20phosphorylates%20Endosulfine%20and%20Arpp19%20to%20convert%20them%20into%20inhibitors%20of%20PP2A-B55%20phosphatase%2C%20thereby%20preventing%20early%20dephosphorylation%20of%20M-phase%20targets%20of%20Cyclin%20B-Cdk1.%20Here%2C%20searching%20for%20an%20elusive%20Gwl-like%20activity%20in%20C.%20elegans%2C%20we%20show%20that%20the%20single%20worm%20MAST%20kinase%2C%20KIN-4%2C%20fulfills%20this%20function%20in%20worms%20and%20can%20functionally%20replace%20Greatwall%20in%20the%20heterologous%20Xenopus%20system.%20Compared%20to%20Greatwall%2C%20the%20short%20activation%20segment%20of%20KIN-4%20lacks%20a%20phosphorylation%20site%2C%20and%20KIN-4%20is%20active%20even%20when%20produced%20in%20E.%20coli.%20We%20also%20show%20that%20a%20balance%20between%20Cyclin%20B-Cdk1%20and%20PP2A-B55%20activity%2C%20regulated%20by%20KIN-4%2C%20is%20essential%20to%20ensure%20asynchronous%20cell%20divisions%20in%20the%20early%20worm%20embryo.%20These%20findings%20resolve%20a%20long-standing%20puzzle%20related%20to%20the%20supposed%20absence%20of%20a%20Greatwall%20pathway%20in%20C.%20elegans%2C%20and%20highlight%20a%20novel%20aspect%20of%20PP2A-B55%20regulation%20by%20MAST%20kinases.%22%2C%22date%22%3A%222025-02-17%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs44318-025-00364-w%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.1038%5C%2Fs44318-025-00364-w%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220261-4189%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222025-02-18T09%3A31%3A22Z%22%7D%7D%2C%7B%22key%22%3A%22BQELGKWL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22El%20Mossadeq%20et%20al.%22%2C%22parsedDate%22%3A%222024-12-26%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BEl%20Mossadeq%2C%20L.%2C%20Bellutti%2C%20L.%2C%20Le%20Borgne%2C%20R.%2C%20Canman%2C%20J.%20C.%2C%20Pintard%2C%20L.%2C%20Verbavatz%2C%20J.-M.%2C%20Askjaer%2C%20P.%2C%20%26amp%3B%20Dumont%2C%20J.%20%282024%29.%20An%20interkinetic%20envelope%20surrounds%20chromosomes%20between%20meiosis%20I%20and%20II%20in%20C.%20elegans%20oocytes.%20%26lt%3Bi%26gt%3BJournal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B224%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20e202403125.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202403125%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202403125%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22An%20interkinetic%20envelope%20surrounds%20chromosomes%20between%20meiosis%20I%20and%20II%20in%20C.%20elegans%20oocytes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Layla%22%2C%22lastName%22%3A%22El%20Mossadeq%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Bellutti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00e9mi%22%2C%22lastName%22%3A%22Le%20Borgne%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julie%20C.%22%2C%22lastName%22%3A%22Canman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Marc%22%2C%22lastName%22%3A%22Verbavatz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%22%2C%22lastName%22%3A%22Askjaer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Dumont%22%7D%5D%2C%22abstractNote%22%3A%22At%20the%20end%20of%20cell%20division%2C%20the%20nuclear%20envelope%20reassembles%20around%20the%20decondensing%20chromosomes.%20Female%20meiosis%20culminates%20in%20two%20consecutive%20cell%20divisions%20of%20the%20oocyte%2C%20meiosis%20I%20and%20II%2C%20which%20are%20separated%20by%20a%20brief%20transition%20phase%20known%20as%20interkinesis.%20Due%20to%20the%20absence%20of%20chromosome%20decondensation%20and%20the%20suppression%20of%20genome%20replication%20during%20interkinesis%2C%20it%20has%20been%20widely%20assumed%20that%20the%20nuclear%20envelope%20does%20not%20reassemble%20between%20meiosis%20I%20and%20II.%20By%20analyzing%20interkinesis%20in%20C.%20elegans%20oocytes%2C%20we%20instead%20show%20that%20an%20atypical%20structure%20made%20of%20two%20lipid%20bilayers%2C%20which%20we%20termed%20the%20interkinetic%20envelope%2C%20surrounds%20the%20surface%20of%20the%20segregating%20chromosomes.%20The%20interkinetic%20envelope%20shares%20common%20features%20with%20the%20nuclear%20envelope%20but%20also%20exhibits%20specific%20characteristics%20that%20distinguish%20it%2C%20including%20its%20lack%20of%20continuity%20with%20the%20endoplasmic%20reticulum%2C%20unique%20protein%20composition%2C%20assembly%20mechanism%2C%20and%20function%20in%20chromosome%20segregation.%20These%20distinct%20attributes%20collectively%20define%20the%20interkinetic%20envelope%20as%20a%20unique%20and%20specialized%20structure%20that%20has%20been%20previously%20overlooked.%22%2C%22date%22%3A%222024-12-26%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202403125%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202403125%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220021-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22I7CUV6U5%22%2C%22MH4QXJHX%22%2C%22GWSKYQWE%22%2C%227DC6IIUA%22%5D%2C%22dateModified%22%3A%222025-01-02T10%3A15%3A31Z%22%7D%7D%2C%7B%22key%22%3A%22Z4276ISH%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Strzelecki%20et%20al.%22%2C%22parsedDate%22%3A%222024-09-28%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BStrzelecki%2C%20P.%2C%20Joly%2C%20N.%2C%20H%26%23xE9%3Bbraud%2C%20P.%2C%20Hoffmann%2C%20E.%2C%20Cech%2C%20G.%20M.%2C%20Kloska%2C%20A.%2C%20Busi%2C%20F.%2C%20%26amp%3B%20Grange%2C%20W.%20%282024%29.%20Enhanced%20Golden%20Gate%20Assembly%3A%20evaluating%20overhang%20strength%20for%20improved%20ligation%20efficiency.%20%26lt%3Bi%26gt%3BNucleic%20Acids%20Research%26lt%3B%5C%2Fi%26gt%3B%2C%20gkae809.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkae809%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkae809%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Enhanced%20Golden%20Gate%20Assembly%3A%20evaluating%20overhang%20strength%20for%20improved%20ligation%20efficiency%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patryk%22%2C%22lastName%22%3A%22Strzelecki%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pascal%22%2C%22lastName%22%3A%22H%5Cu00e9braud%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elise%22%2C%22lastName%22%3A%22Hoffmann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Grzegorz%5Cu00a0M%22%2C%22lastName%22%3A%22Cech%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Kloska%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Florent%22%2C%22lastName%22%3A%22Busi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Wilfried%22%2C%22lastName%22%3A%22Grange%22%7D%5D%2C%22abstractNote%22%3A%22Molecular%20cloning%2C%20a%20routine%20yet%20essential%20technique%2C%20relies%20heavily%20on%20efficient%20ligation%2C%20which%20can%20be%20significantly%20improved%20using%20Golden%20Gate%5Cu00a0Assembly%20%28GGA%29.%20A%20key%20component%20of%20GGA%20is%20the%20use%20of%20type%20IIS%20enzymes%2C%20which%20uniquely%20cleave%20downstream%20of%20their%20recognition%20sequences%20to%20generate%20various%20overhangs%2C%20including%20non-palindromic%20ones.%20Recent%20advancements%20in%20GGA%20include%20the%20development%20of%20newly%20engineered%20enzymes%20with%20enhanced%20activity.%20Additionally%2C%20high-throughput%20GGA%20assays%2C%20which%20allow%20for%20the%20simultaneous%20study%20of%20all%20possible%20overhangs%2C%20have%20identified%20optimal%20GGA%20substrates%20with%20high%20efficiencies%20and%20fidelities%2C%20greatly%20facilitating%20the%20design%20of%20complex%20assemblies.%20Interestingly%2C%20these%20assays%20reveal%20unexpected%20correlations%20between%20ligation%20efficiencies%20and%20overhang%20stabilities.%20One%20hypothesis%20for%20this%20observation%20is%20that%20newly%20hydrolyzed%20DNA%20fragments%20with%20strong%20overhangs%20can%20readily%20re-ligate%2C%20thereby%20slowing%20down%20the%20overall%20process.%20In%20this%20paper%2C%20we%20employ%20a%20combination%20of%20gel%20electrophoresis%20and%20numerical%20calculations%20to%20test%20this%20hypothesis%2C%20ultimately%20determining%20that%20it%20does%20not%20hold%20true%20under%20the%20conditions%20established%20by%20conventional%20GGA%20assays.%20Using%20an%20assembly%20of%2010%20fragments%2C%20we%20demonstrate%20that%20strong%20overhangs%20yield%20higher%20GGA%20efficiency%2C%20while%20weak%20overhangs%20result%20in%20lower%20efficiency.%20These%20findings%20enable%20us%20to%20propose%20optimal%20overhangs%20for%20efficient%20GGA%20assays%2C%20significantly%20increasing%20yield.%22%2C%22date%22%3A%222024-09-28%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkae809%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkae809%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220305-1048%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222024-09-30T08%3A55%3A27Z%22%7D%7D%2C%7B%22key%22%3A%229YPAUBFG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Beaumale%20et%20al.%22%2C%22parsedDate%22%3A%222024-02-08%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBeaumale%2C%20E.%2C%20Van%20Hove%2C%20L.%2C%20Pintard%2C%20L.%2C%20%26amp%3B%20Joly%2C%20N.%20%282024%29.%20Microtubule-binding%20domains%20in%20Katanin%20p80%20subunit%20are%20essential%20for%20severing%20activity%20in%20C.%20elegans.%20%26lt%3Bi%26gt%3BJournal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B223%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20e202308023.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202308023%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202308023%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Microtubule-binding%20domains%20in%20Katanin%20p80%20subunit%20are%20essential%20for%20severing%20activity%20in%20C.%20elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eva%22%2C%22lastName%22%3A%22Beaumale%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%5D%2C%22abstractNote%22%3A%22Microtubule-severing%20enzymes%20%28MSEs%29%2C%20such%20as%20Katanin%2C%20Spastin%2C%20and%20Fidgetin%20play%20essential%20roles%20in%20cell%20division%20and%20neurogenesis.%20They%20damage%20the%20microtubule%20%28MT%29%20lattice%2C%20which%20can%20either%20destroy%20or%20amplify%20the%20MT%20cytoskeleton%2C%20depending%20on%20the%20cellular%20context.%20However%2C%20little%20is%20known%20about%20how%20they%20interact%20with%20their%20substrates.%20We%20have%20identified%20the%20microtubule-binding%20domains%20%28MTBD%29%20required%20for%20Katanin%20function%20in%20C.%20elegans.%20Katanin%20is%20a%20heterohexamer%20of%20dimers%20containing%20a%20catalytic%20subunit%20p60%20and%20a%20regulatory%20subunit%20p80%2C%20both%20of%20which%20are%20essential%20for%20female%20meiotic%20spindle%20assembly.%20Here%2C%20we%20report%20that%20p80-like%28MEI-2%29%20dictates%20Katanin%20binding%20to%20MTs%20via%20two%20MTBDs%20composed%20of%20basic%20patches.%20Substituting%20these%20patches%20reduces%20Katanin%20binding%20to%20MTs%2C%20compromising%20its%20function%20in%20female%20meiotic-spindle%20assembly.%20Structural%20alignments%20of%20p80-like%28MEI-2%29%20with%20p80s%20from%20different%20species%20revealed%20that%20the%20MTBDs%20are%20evolutionarily%20conserved%2C%20even%20if%20the%20specific%20amino%20acids%20involved%20vary.%20Our%20findings%20highlight%20the%20critical%20importance%20of%20the%20regulatory%20subunit%20%28p80%29%20in%20providing%20MT%20binding%20to%20the%20Katanin%20complex.%22%2C%22date%22%3A%222024-02-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202308023%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202308023%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220021-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222024-02-13T08%3A46%3A43Z%22%7D%7D%2C%7B%22key%22%3A%22JLH8QS2U%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Velez-Aguilera%20et%20al.%22%2C%22parsedDate%22%3A%222024%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVelez-Aguilera%2C%20G.%2C%20Ossareh-Nazari%2C%20B.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282024%29.%20Dissecting%20the%20Multiple%20Functions%20of%20the%20Polo-Like%20Kinase%201%20in%20the%20C.%20elegans%20Zygote.%20In%20A.%20Castro%20%26amp%3B%20B.%20Lacroix%20%28Eds.%29%2C%20%26lt%3Bi%26gt%3BCell%20Cycle%20Control%3A%20Methods%20and%20Protocols%26lt%3B%5C%2Fi%26gt%3B%20%28pp.%2063%26%23×2013%3B88%29.%20Springer%20US.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-1-0716-3557-5_4%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-1-0716-3557-5_4%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22bookSection%22%2C%22title%22%3A%22Dissecting%20the%20Multiple%20Functions%20of%20the%20Polo-Like%20Kinase%201%20in%20the%20C.%20elegans%20Zygote%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Benjamin%22%2C%22lastName%22%3A%22Lacroix%22%7D%5D%2C%22abstractNote%22%3A%22Plk1%20%28polo-like%20kinase%201%29%20is%20an%20evolutionarily%20conserved%20serine%5C%2Fthreonine%20kinase%20instrumental%20for%20mitotic%20entry%20and%20progression.%20Beyond%20these%20canonical%20functions%2C%20Plk1%20also%20regulates%20cell%20polarization%20and%20cell%20fate%20during%20asymmetric%20cell%20divisions%20in%20C.%20elegans%20and%20D.%20melanogaster.%20Plk1%20contains%20a%20specialized%20phosphoserine%5Cu2013threonine%20binding%20domain%2C%20the%20polo-box%20domain%20%28PBD%29%2C%20which%20localizes%20and%20concentrates%20the%20kinase%20at%20its%20various%20sites%20of%20action%20within%20the%20cell%20in%20space%20and%20time.%20Here%20we%20present%20protocols%20to%20express%20and%20purify%20the%20C.%20elegans%20Plk1%20kinase%20along%20with%20biochemical%20and%20phosphoproteomic%20approaches%20to%20interrogate%20the%20PBD%20interactome%20and%20to%20dissect%20Plk1%20substrate%20interactions.%20These%20protocols%20are%20most%20suitable%20for%20the%20identification%20of%20Plk1%20targets%20in%20C.%20elegans%20embryos%20but%20can%20be%20easily%20adapted%20to%20identify%20and%20study%20Plk1%20substrates%20from%20any%20source.%5Cu201d%22%2C%22bookTitle%22%3A%22Cell%20Cycle%20Control%3A%20Methods%20and%20Protocols%22%2C%22date%22%3A%222024%22%2C%22originalDate%22%3A%22%22%2C%22originalPublisher%22%3A%22%22%2C%22originalPlace%22%3A%22%22%2C%22format%22%3A%22%22%2C%22ISBN%22%3A%22978-1-07-163557-5%22%2C%22DOI%22%3A%22%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-1-0716-3557-5_4%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222024-02-27T09%3A30%3A16Z%22%7D%7D%2C%7B%22key%22%3A%22JMRE4R57%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Nkombo%20Nkoula%20et%20al.%22%2C%22parsedDate%22%3A%222023-07-19%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BNkombo%20Nkoula%2C%20S.%2C%20Velez-Aguilera%2C%20G.%2C%20Ossareh-Nazari%2C%20B.%2C%20Van%20Hove%2C%20L.%2C%20Ayuso%2C%20C.%2C%20Legros%2C%20V.%2C%20Chevreux%2C%20G.%2C%20Thomas%2C%20L.%2C%20Seydoux%2C%20G.%2C%20Askjaer%2C%20P.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282023%29.%20Mechanisms%20of%20nuclear%20pore%20complex%20disassembly%20by%20the%20mitotic%20Polo-like%20kinase%201%20%28PLK-1%29%20in%20C.%20elegans%20embryos.%20%26lt%3Bi%26gt%3BScience%20Advances%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2829%29%2C%20eadf7826.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fsciadv.adf7826%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fsciadv.adf7826%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Mechanisms%20of%20nuclear%20pore%20complex%20disassembly%20by%20the%20mitotic%20Polo-like%20kinase%201%20%28PLK-1%29%20in%20C.%20elegans%20embryos%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sylvia%22%2C%22lastName%22%3A%22Nkombo%20Nkoula%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cristina%22%2C%22lastName%22%3A%22Ayuso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22V%5Cu00e9ronique%22%2C%22lastName%22%3A%22Legros%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Guillaume%22%2C%22lastName%22%3A%22Chevreux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Thomas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G%5Cu00e9raldine%22%2C%22lastName%22%3A%22Seydoux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%22%2C%22lastName%22%3A%22Askjaer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22The%20nuclear%20envelope%2C%20which%20protects%20and%20organizes%20the%20genome%2C%20is%20dismantled%20during%20mitosis.%20In%20the%20Caenorhabditis%20elegans%20zygote%2C%20nuclear%20envelope%20breakdown%20%28NEBD%29%20of%20the%20parental%20pronuclei%20is%20spatially%20and%20temporally%20regulated%20during%20mitosis%20to%20promote%20the%20unification%20of%20the%20maternal%20and%20paternal%20genomes.%20Nuclear%20pore%20complex%20%28NPC%29%20disassembly%20is%20a%20decisive%20step%20of%20NEBD%2C%20essential%20for%20nuclear%20permeabilization.%20By%20combining%20live%20imaging%2C%20biochemistry%2C%20and%20phosphoproteomics%2C%20we%20show%20that%20NPC%20disassembly%20is%20a%20stepwise%20process%20that%20involves%20Polo-like%20kinase%201%20%28PLK-1%29%5Cu2013dependent%20and%20%5Cu2013independent%20steps.%20PLK-1%20targets%20multiple%20NPC%20subcomplexes%2C%20including%20the%20cytoplasmic%20filaments%2C%20central%20channel%2C%20and%20inner%20ring.%20PLK-1%20is%20recruited%20to%20and%20phosphorylates%20intrinsically%20disordered%20regions%20%28IDRs%29%20of%20several%20multivalent%20linker%20nucleoporins.%20Notably%2C%20although%20the%20phosphosites%20are%20not%20conserved%20between%20human%20and%20C.%20elegans%20nucleoporins%2C%20they%20are%20located%20in%20IDRs%20in%20both%20species.%20Our%20results%20suggest%20that%20targeting%20IDRs%20of%20multivalent%20linker%20nucleoporins%20is%20an%20evolutionarily%20conserved%20driver%20of%20NPC%20disassembly%20during%20mitosis.%22%2C%22date%22%3A%222023-07-19%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1126%5C%2Fsciadv.adf7826%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.science.org%5C%2Fdoi%5C%2F10.1126%5C%2Fsciadv.adf7826%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222023-07-21T12%3A26%3A04Z%22%7D%7D%2C%7B%22key%22%3A%227SJ3YXZD%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Kouranti%20et%20al.%22%2C%22parsedDate%22%3A%222022-05-05%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BKouranti%2C%20I.%2C%20Abdel%20Khalek%2C%20W.%2C%20Mazurkiewicz%2C%20S.%2C%20Loisel-Ferreira%2C%20I.%2C%20Gautreau%2C%20A.%20M.%2C%20Pintard%2C%20L.%2C%20Jeunemaitre%2C%20X.%2C%20%26amp%3B%20Clauser%2C%20E.%20%282022%29.%20Cullin%203%20Exon%209%20Deletion%20in%20Familial%20Hyperkalemic%20Hypertension%20Impairs%20Cullin3-Ring-E3%20Ligase%20%28CRL3%29%20Dynamic%20Regulation%20and%20Cycling.%20%26lt%3Bi%26gt%3BInternational%20Journal%20of%20Molecular%20Sciences%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B23%26lt%3B%5C%2Fi%26gt%3B%289%29%2C%205151.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms23095151%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms23095151%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Cullin%203%20Exon%209%20Deletion%20in%20Familial%20Hyperkalemic%20Hypertension%20Impairs%20Cullin3-Ring-E3%20Ligase%20%28CRL3%29%20Dynamic%20Regulation%20and%20Cycling%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ilektra%22%2C%22lastName%22%3A%22Kouranti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Waed%22%2C%22lastName%22%3A%22Abdel%20Khalek%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stephani%22%2C%22lastName%22%3A%22Mazurkiewicz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Irmine%22%2C%22lastName%22%3A%22Loisel-Ferreira%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alexis%20M.%22%2C%22lastName%22%3A%22Gautreau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xavier%22%2C%22lastName%22%3A%22Jeunemaitre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%22%2C%22lastName%22%3A%22Clauser%22%7D%5D%2C%22abstractNote%22%3A%22Cullin%203%20%28CUL3%29%20is%20the%20scaffold%20of%20Cullin3%20Ring%20E3-ligases%20%28CRL3s%29%2C%20which%20use%20various%20BTB-adaptor%20proteins%20to%20ubiquitinate%20numerous%20substrates%20targeting%20their%20proteasomal%20degradation.%20CUL3%20mutations%2C%20responsible%20for%20a%20severe%20form%20of%20familial%20hyperkalemia%20and%20hypertension%20%28FHHt%29%2C%20all%20result%20in%20a%20deletion%20of%20exon%209%20%28amino-acids%20403-459%29%20%28CUL3-%5Cu22069%29.%20Surprisingly%2C%20while%20CUL3-%5Cu22069%20is%20hyperneddylated%2C%20a%20post-translational%20modification%20that%20typically%20activates%20CRL%20complexes%2C%20it%20is%20unable%20to%20ubiquitinate%20its%20substrates.%20In%20order%20to%20understand%20the%20mechanisms%20behind%20this%20loss-of%20function%2C%20we%20performed%20comparative%20label-free%20quantitative%20analyses%20of%20CUL3%20and%20CUL3-%5Cu22069%20interactome%20by%20mass%20spectrometry.%20It%20was%20observed%20that%20CUL3-%5Cu22069%20interactions%20with%20COP9%20and%20CAND1%2C%20both%20involved%20in%20CRL3%20complexes%26%23039%3B%20dynamic%20assembly%2C%20were%20disrupted.%20These%20defects%20result%20in%20a%20reduction%20in%20the%20dynamic%20cycling%20of%20the%20CRL3%20complexes%2C%20making%20the%20CRL3-%5Cu22069%20complex%20an%20inactive%20BTB-adaptor%20trap%2C%20as%20demonstrated%20by%20SILAC%20experiments.%20Collectively%2C%20the%20data%20indicated%20that%20the%20hyperneddylated%20CUL3-%5Cu22069%20protein%20is%20inactive%20as%20a%20consequence%20of%20several%20structural%20changes%20disrupting%20its%20dynamic%20interactions%20with%20key%20regulatory%20partners.%22%2C%22date%22%3A%222022-05-05%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.3390%5C%2Fijms23095151%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221422-0067%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A25%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22ZJWMJGLH%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Pillan%20et%20al.%22%2C%22parsedDate%22%3A%222022-04%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPillan%2C%20A.%2C%20Tavernier%2C%20N.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282022%29.%20%5BThe%20kiss%20of%20life%3A%20Aurora%20A%20embraces%20the%20phosphate%20of%20its%20cofactor%20Bora%20to%20trigger%20mitotic%20entry%5D.%20%26lt%3Bi%26gt%3BMedecine%20Sciences%3A%20M%5C%2FS%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B38%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20345%26%23×2013%3B347.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1051%5C%2Fmedsci%5C%2F2022033%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1051%5C%2Fmedsci%5C%2F2022033%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22%5BThe%20kiss%20of%20life%3A%20Aurora%20A%20embraces%20the%20phosphate%20of%20its%20cofactor%20Bora%20to%20trigger%20mitotic%20entry%5D%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ana%5Cu00efs%22%2C%22lastName%22%3A%22Pillan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222022-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1051%5C%2Fmedsci%5C%2F2022033%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221958-5381%22%2C%22language%22%3A%22fre%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A25%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22GI4PXUMF%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Velez-Aguilera%20et%20al.%22%2C%22parsedDate%22%3A%222022-03-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVelez-Aguilera%2C%20G.%2C%20Ossareh-Nazari%2C%20B.%2C%20Van%20Hove%2C%20L.%2C%20Joly%2C%20N.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282022%29.%20Cortical%20microtubule%20pulling%20forces%20contribute%20to%20the%20union%20of%20the%20parental%20genomes%20in%20the%20Caenorhabditis%20elegans%20zygote.%20%26lt%3Bi%26gt%3BeLife%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B11%26lt%3B%5C%2Fi%26gt%3B%2C%20e75382.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.75382%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.75382%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Cortical%20microtubule%20pulling%20forces%20contribute%20to%20the%20union%20of%20the%20parental%20genomes%20in%20the%20Caenorhabditis%20elegans%20zygote%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Previously%2C%20we%20reported%20that%20the%20Polo-like%20kinase%20PLK-1%20phosphorylates%20the%20single%20Caenorhabditis%20elegans%20lamin%20%28LMN-1%29%20to%20trigger%20lamina%20depolymerization%20during%20mitosis.%20We%20showed%20that%20this%20event%20is%20required%20to%20form%20a%20pronuclear%20envelope%20scission%20event%20that%20removes%20membranes%20on%20the%20juxtaposed%20oocyte%20and%20sperm%20pronuclear%20envelopes%20in%20the%20zygote%2C%20allowing%20the%20parental%20chromosomes%20to%20merge%20in%20a%20single%20nucleus%20after%20segregation%20%28Velez-Aguilera%20et%20al.%2C%202020%29.%20Here%2C%20we%20show%20that%20cortical%20microtubule%20pulling%20forces%20contribute%20to%20pronuclear%20envelopes%20scission%20by%20promoting%20mitotic%20spindle%20elongation%2C%20and%20conversely%2C%20nuclear%20envelopes%20remodeling%20facilitates%20spindle%20elongation.%20We%20also%20demonstrate%20that%20weakening%20the%20pronuclear%20envelopes%20via%20PLK-1-mediated%20lamina%20depolymerization%2C%20is%20a%20prerequisite%20for%20the%20astral%20microtubule%20pulling%20forces%20to%20trigger%20pronuclear%20membranes%20scission.%20Finally%2C%20we%20provide%20evidence%20that%20PLK-1%20mainly%20acts%20via%20lamina%20depolymerization%20in%20this%20process.%20These%20observations%20thus%20indicate%20that%20temporal%20coordination%20between%20lamina%20depolymerization%20and%20mitotic%20spindle%20elongation%20facilitates%20pronuclear%20envelopes%20scission%20and%20parental%20genomes%20unification.%22%2C%22date%22%3A%222022-03-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.75382%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222050-084X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22BXVTCIE8%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Tavernier%20et%20al.%22%2C%22parsedDate%22%3A%222021-09-06%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BTavernier%2C%20N.%2C%20Sicheri%2C%20F.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282021%29.%20Aurora%20A%20kinase%20activation%3A%20Different%20means%20to%20different%20ends.%20%26lt%3Bi%26gt%3BThe%20Journal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B220%26lt%3B%5C%2Fi%26gt%3B%289%29%2C%20e202106128.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202106128%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202106128%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Aurora%20A%20kinase%20activation%3A%20Different%20means%20to%20different%20ends%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Frank%22%2C%22lastName%22%3A%22Sicheri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Aurora%20A%20is%20a%20serine%5C%2Fthreonine%20kinase%20essential%20for%20mitotic%20entry%20and%20spindle%20assembly.%20Recent%20molecular%20studies%20have%20revealed%20the%20existence%20of%20multiple%2C%20distinct%20mechanisms%20of%20Aurora%20A%20activation%2C%20each%20occurring%20at%20specific%20subcellular%20locations%2C%20optimized%20for%20cellular%20context%2C%20and%20primed%20by%20signaling%20events%20including%20phosphorylation%20and%20oxidation.%22%2C%22date%22%3A%222021-09-06%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202106128%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221540-8140%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22R2DYQDHX%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Knox%20et%20al.%22%2C%22parsedDate%22%3A%222021-04-28%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BKnox%2C%20J.%2C%20Joly%2C%20N.%2C%20Linossi%2C%20E.%20M.%2C%20Carmona-Negr%26%23xF3%3Bn%2C%20J.%20A.%2C%20Jura%2C%20N.%2C%20Pintard%2C%20L.%2C%20Zuercher%2C%20W.%2C%20%26amp%3B%20Roy%2C%20P.%20J.%20%282021%29.%20A%20survey%20of%20the%20kinome%20pharmacopeia%20reveals%20multiple%20scaffolds%20and%20targets%20for%20the%20development%20of%20novel%20anthelmintics.%20%26lt%3Bi%26gt%3BScientific%20Reports%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B11%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%209161.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-021-88150-6%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-021-88150-6%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20survey%20of%20the%20kinome%20pharmacopeia%20reveals%20multiple%20scaffolds%20and%20targets%20for%20the%20development%20of%20novel%20anthelmintics%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%22%2C%22lastName%22%3A%22Knox%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Edmond%20M.%22%2C%22lastName%22%3A%22Linossi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jos%5Cu00e9%20A.%22%2C%22lastName%22%3A%22Carmona-Negr%5Cu00f3n%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Natalia%22%2C%22lastName%22%3A%22Jura%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22William%22%2C%22lastName%22%3A%22Zuercher%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20J.%22%2C%22lastName%22%3A%22Roy%22%7D%5D%2C%22abstractNote%22%3A%22Over%20one%20billion%20people%20are%20currently%20infected%20with%20a%20parasitic%20nematode.%20Symptoms%20can%20include%20anemia%2C%20malnutrition%2C%20developmental%20delay%2C%20and%20in%20severe%20cases%2C%20death.%20Resistance%20is%20emerging%20to%20the%20anthelmintics%20currently%20used%20to%20treat%20nematode%20infection%2C%20prompting%20the%20need%20to%20develop%20new%20anthelmintics.%20Towards%20this%20end%2C%20we%20identified%20a%20set%20of%20kinases%20that%20may%20be%20targeted%20in%20a%20nematode-selective%20manner.%20We%20first%20screened%202040%20inhibitors%20of%20vertebrate%20kinases%20for%20those%20that%20impair%20the%20model%20nematode%20Caenorhabditis%20elegans.%20By%20determining%20whether%20the%20terminal%20phenotype%20induced%20by%20each%20kinase%20inhibitor%20matched%20that%20of%20the%20predicted%20target%20mutant%20in%20C.%20elegans%2C%20we%20identified%2017%20druggable%20nematode%20kinase%20targets.%20Of%20these%2C%20we%20found%20that%20nematode%20EGFR%2C%20MEK1%2C%20and%20PLK1%20kinases%20have%20diverged%20from%20vertebrates%20within%20their%20drug-binding%20pocket.%20For%20each%20of%20these%20targets%2C%20we%20identified%20small%20molecule%20scaffolds%20that%20may%20be%20further%20modified%20to%20develop%20nematode-selective%20inhibitors.%20Nematode%20EGFR%2C%20MEK1%2C%20and%20PLK1%20therefore%20represent%20key%20targets%20for%20the%20development%20of%20new%20anthelmintic%20medicines.%22%2C%22date%22%3A%222021-04-28%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41598-021-88150-6%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222045-2322%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%2287GI54GG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Tavernier%20et%20al.%22%2C%22parsedDate%22%3A%222021-03-26%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BTavernier%2C%20N.%2C%20Thomas%2C%20Y.%2C%20Vigneron%2C%20S.%2C%20Maisonneuve%2C%20P.%2C%20Orlicky%2C%20S.%2C%20Mader%2C%20P.%2C%20Regmi%2C%20S.%20G.%2C%20Van%20Hove%2C%20L.%2C%20Levinson%2C%20N.%20M.%2C%20Gasmi-Seabrook%2C%20G.%2C%20Joly%2C%20N.%2C%20Poteau%2C%20M.%2C%20Velez-Aguilera%2C%20G.%2C%20Gavet%2C%20O.%2C%20Castro%2C%20A.%2C%20Dasso%2C%20M.%2C%20Lorca%2C%20T.%2C%20Sicheri%2C%20F.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282021%29.%20Bora%20phosphorylation%20substitutes%20in%20trans%20for%20T-loop%20phosphorylation%20in%20Aurora%20A%20to%20promote%20mitotic%20entry.%20%26lt%3Bi%26gt%3BNature%20Communications%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B12%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%201899.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-021-21922-w%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-021-21922-w%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Bora%20phosphorylation%20substitutes%20in%20trans%20for%20T-loop%20phosphorylation%20in%20Aurora%20A%20to%20promote%20mitotic%20entry%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y.%22%2C%22lastName%22%3A%22Thomas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%22%2C%22lastName%22%3A%22Vigneron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22P.%22%2C%22lastName%22%3A%22Maisonneuve%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%22%2C%22lastName%22%3A%22Orlicky%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22P.%22%2C%22lastName%22%3A%22Mader%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20G.%22%2C%22lastName%22%3A%22Regmi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%20M.%22%2C%22lastName%22%3A%22Levinson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G.%22%2C%22lastName%22%3A%22Gasmi-Seabrook%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Poteau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G.%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22O.%22%2C%22lastName%22%3A%22Gavet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22A.%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Dasso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22T.%22%2C%22lastName%22%3A%22Lorca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22F.%22%2C%22lastName%22%3A%22Sicheri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Polo-like%20kinase%201%20%28Plk1%29%20is%20instrumental%20for%20mitotic%20entry%20and%20progression.%20Plk1%20is%20activated%20by%20phosphorylation%20on%20a%20conserved%20residue%20Thr210%20in%20its%20activation%20segment%20by%20the%20Aurora%20A%20kinase%20%28AURKA%29%2C%20a%20reaction%20that%20critically%20requires%20the%20co-factor%20Bora%20phosphorylated%20by%20a%20CyclinA%5C%2FB-Cdk1%20kinase.%20Here%20we%20show%20that%20phospho-Bora%20is%20a%20direct%20activator%20of%20AURKA%20kinase%20activity.%20We%20localize%20the%20key%20determinants%20of%20phospho-Bora%20function%20to%20a%20100%20amino%20acid%20region%20encompassing%20two%20short%20Tpx2-like%20motifs%20and%20a%20phosphoSerine-Proline%20motif%20at%20Serine%20112%2C%20through%20which%20Bora%20binds%20AURKA.%20The%20latter%20substitutes%20in%20trans%20for%20the%20Thr288%20phospho-regulatory%20site%20of%20AURKA%2C%20which%20is%20essential%20for%20an%20active%20conformation%20of%20the%20kinase%20domain.%20We%20demonstrate%20the%20importance%20of%20these%20determinants%20for%20Bora%20function%20in%20mitotic%20entry%20both%20in%20Xenopus%20egg%20extracts%20and%20in%5Cu00a0human%20cells.%20Our%20findings%20unveil%20the%20activation%20mechanism%20of%20AURKA%20that%20is%20critical%20for%20mitotic%20entry.%22%2C%22date%22%3A%222021-03-26%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41467-021-21922-w%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222041-1723%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22JDPEX3E3%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Velez-Aguilera%20et%20al.%22%2C%22parsedDate%22%3A%222020-10-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVelez-Aguilera%2C%20G.%2C%20Nkombo%20Nkoula%2C%20S.%2C%20Ossareh-Nazari%2C%20B.%2C%20Link%2C%20J.%2C%20Paouneskou%2C%20D.%2C%20Van%20Hove%2C%20L.%2C%20Joly%2C%20N.%2C%20Tavernier%2C%20N.%2C%20Verbavatz%2C%20J.-M.%2C%20Jantsch%2C%20V.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282020%29.%20PLK-1%20promotes%20the%20merger%20of%20the%20parental%20genome%20into%20a%20single%20nucleus%20by%20triggering%20lamina%20disassembly.%20%26lt%3Bi%26gt%3BeLife%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2C%20e59510.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.59510%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.59510%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22PLK-1%20promotes%20the%20merger%20of%20the%20parental%20genome%20into%20a%20single%20nucleus%20by%20triggering%20lamina%20disassembly%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sylvia%22%2C%22lastName%22%3A%22Nkombo%20Nkoula%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jana%22%2C%22lastName%22%3A%22Link%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dimitra%22%2C%22lastName%22%3A%22Paouneskou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Marc%22%2C%22lastName%22%3A%22Verbavatz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Verena%22%2C%22lastName%22%3A%22Jantsch%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Life%20of%20sexually%20reproducing%20organisms%20starts%20with%20the%20fusion%20of%20the%20haploid%20egg%20and%20sperm%20gametes%20to%20form%20the%20genome%20of%20a%20new%20diploid%20organism.%20Using%20the%20newly%20fertilized%20Caenorhabditis%20elegans%20zygote%2C%20we%20show%20that%20the%20mitotic%20Polo-like%20kinase%20PLK-1%20phosphorylates%20the%20lamin%20LMN-1%20to%20promote%20timely%20lamina%20disassembly%20and%20subsequent%20merging%20of%20the%20parental%20genomes%20into%20a%20single%20nucleus%20after%20mitosis.%20Expression%20of%20non-phosphorylatable%20versions%20of%20LMN-1%2C%20which%20affect%20lamina%20depolymerization%20during%20mitosis%2C%20is%20sufficient%20to%20prevent%20the%20mixing%20of%20the%20parental%20chromosomes%20into%20a%20single%20nucleus%20in%20daughter%20cells.%20Finally%2C%20we%20recapitulate%20lamina%20depolymerization%20by%20PLK-1%20in%20vitro%20demonstrating%20that%20LMN-1%20is%20a%20direct%20PLK-1%20target.%20Our%20findings%20indicate%20that%20the%20timely%20removal%20of%20lamin%20is%20essential%20for%20the%20merging%20of%20parental%20chromosomes%20at%20the%20beginning%20of%20life%20in%20C.%20elegans%20and%20possibly%20also%20in%20humans%2C%20where%20a%20defect%20in%20this%20process%20might%20be%20fatal%20for%20embryo%20development.%22%2C%22date%22%3A%222020-10-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.59510%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222050-084X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%229J76DXBR%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Joly%20et%20al.%22%2C%22parsedDate%22%3A%222020-06-01%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BJoly%2C%20N.%2C%20Beaumale%2C%20E.%2C%20Van%20Hove%2C%20L.%2C%20Martino%2C%20L.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282020%29.%20Phosphorylation%20of%20the%20microtubule-severing%20AAA%2B%20enzyme%20Katanin%20regulates%20C.%20elegans%20embryo%20development.%20%26lt%3Bi%26gt%3BThe%20Journal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B219%26lt%3B%5C%2Fi%26gt%3B%286%29%2C%20e201912037.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.201912037%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.201912037%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Phosphorylation%20of%20the%20microtubule-severing%20AAA%2B%20enzyme%20Katanin%20regulates%20C.%20elegans%20embryo%20development%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eva%22%2C%22lastName%22%3A%22Beaumale%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lisa%22%2C%22lastName%22%3A%22Martino%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22The%20evolutionarily%20conserved%20microtubule%20%28MT%29-severing%20AAA-ATPase%20enzyme%20Katanin%20is%20emerging%20as%20a%20critical%20regulator%20of%20MT%20dynamics.%20In%20Caenorhabditis%20elegans%2C%20Katanin%20MT-severing%20activity%20is%20essential%20for%20meiotic%20spindle%20assembly%20but%20is%20toxic%20for%20the%20mitotic%20spindle.%20Here%20we%20analyzed%20Katanin%20dynamics%20in%20C.%20elegans%20and%20deciphered%20the%20role%20of%20Katanin%20phosphorylation%20in%20the%20regulation%20of%20its%20activity%20and%20stability.%20Katanin%20is%20abundant%20in%20oocytes%2C%20and%20its%20levels%20drop%20after%20meiosis%2C%20but%20unexpectedly%2C%20a%20significant%20fraction%20is%20present%20throughout%20embryogenesis%2C%20where%20it%20is%20dynamically%20recruited%20to%20the%20centrosomes%20and%20chromosomes%20during%20mitosis.%20We%20show%20that%20the%20minibrain%20kinase%20MBK-2%2C%20which%20is%20activated%20during%20meiosis%2C%20phosphorylates%20Katanin%20at%20multiple%20serines.%20We%20demonstrate%20unequivocally%20that%20Katanin%20phosphorylation%20at%20a%20single%20residue%20is%20necessary%20and%20sufficient%20to%20target%20Katanin%20for%20proteasomal%20degradation%20after%20meiosis%2C%20whereas%20phosphorylation%20at%20the%20other%20sites%20only%20inhibits%20Katanin%20ATPase%20activity%20stimulated%20by%20MTs.%20Our%20findings%20suggest%20that%20cycles%20of%20phosphorylation%20and%20dephosphorylation%20fine-tune%20Katanin%20level%20and%20activity%20to%20deliver%20the%20appropriate%20MT-severing%20activity%20during%20development.%22%2C%22date%22%3A%222020-06-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.201912037%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221540-8140%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22WSNGXCFC%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Pintard%20and%20Bowerman%22%2C%22parsedDate%22%3A%222019-01%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPintard%2C%20L.%2C%20%26amp%3B%20Bowerman%2C%20B.%20%282019%29.%20Mitotic%20Cell%20Division%20in%20Caenorhabditis%20elegans.%20%26lt%3Bi%26gt%3BGenetics%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B211%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%2035%26%23×2013%3B73.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1534%5C%2Fgenetics.118.301367%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1534%5C%2Fgenetics.118.301367%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Mitotic%20Cell%20Division%20in%20Caenorhabditis%20elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bruce%22%2C%22lastName%22%3A%22Bowerman%22%7D%5D%2C%22abstractNote%22%3A%22Mitotic%20cell%20divisions%20increase%20cell%20number%20while%20faithfully%20distributing%20the%20replicated%20genome%20at%20each%20division.%20The%20Caenorhabditis%20elegans%20embryo%20is%20a%20powerful%20model%20for%20eukaryotic%20cell%20division.%20Nearly%20all%20of%20the%20genes%20that%20regulate%20cell%20division%20in%20C.%20elegans%20are%20conserved%20across%20metazoan%20species%2C%20including%20humans.%20The%20C.%20elegans%20pathways%20tend%20to%20be%20streamlined%2C%20facilitating%20dissection%20of%20the%20more%20redundant%20human%20pathways.%20Here%2C%20we%20summarize%20the%20virtues%20of%20C.%20elegans%20as%20a%20model%20system%20and%20review%20our%20current%20understanding%20of%20centriole%20duplication%2C%20the%20acquisition%20of%20pericentriolar%20material%20by%20centrioles%20to%20form%20centrosomes%2C%20the%20assembly%20of%20kinetochores%20and%20the%20mitotic%20spindle%2C%20chromosome%20segregation%2C%20and%20cytokinesis.%22%2C%22date%22%3A%222019-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1534%5C%2Fgenetics.118.301367%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221943-2631%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%22JTXPFJBL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Pintard%20and%20Archambault%22%2C%22parsedDate%22%3A%222018-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BPintard%2C%20L.%2C%20%26amp%3B%20Archambault%2C%20V.%20%282018%29.%20A%20unified%20view%20of%20spatio-temporal%20control%20of%20mitotic%20entry%3A%20Polo%20kinase%20as%20the%20key.%20%26lt%3Bi%26gt%3BOpen%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B8%26lt%3B%5C%2Fi%26gt%3B%288%29%2C%20180114.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1098%5C%2Frsob.180114%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1098%5C%2Frsob.180114%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20unified%20view%20of%20spatio-temporal%20control%20of%20mitotic%20entry%3A%20Polo%20kinase%20as%20the%20key%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vincent%22%2C%22lastName%22%3A%22Archambault%22%7D%5D%2C%22abstractNote%22%3A%22The%20Polo%20kinase%20is%20an%20essential%20regulator%20of%20cell%20division.%20Its%20ability%20to%20regulate%20multiple%20events%20at%20distinct%20subcellular%20locations%20and%20times%20during%20mitosis%20is%20remarkable.%20In%20the%20last%20few%20years%2C%20a%20much%20clearer%20mechanistic%20understanding%20of%20the%20functions%20and%20regulation%20of%20Polo%20in%20cell%20division%20has%20emerged.%20In%20this%20regard%2C%20the%20importance%20of%20coupling%20changes%20in%20activity%20with%20changes%20in%20localization%20is%20striking%2C%20both%20for%20Polo%20itself%20and%20for%20its%20upstream%20regulators.%20This%20review%20brings%20together%20several%20new%20pieces%20of%20the%20puzzle%20that%20are%20gradually%20revealing%20how%20Polo%20is%20regulated%2C%20in%20space%20and%20time%2C%20to%20enable%20its%20functions%20in%20the%20early%20stages%20of%20mitosis%20in%20animal%20cells.%20As%20a%20result%2C%20a%20unified%20view%20of%20how%20mitotic%20entry%20is%20spatio-temporally%20regulated%20is%20emerging.%22%2C%22date%22%3A%222018-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1098%5C%2Frsob.180114%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222046-2441%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%22IKTDRN4D%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Gutnik%20et%20al.%22%2C%22parsedDate%22%3A%222018-07-16%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BGutnik%2C%20S.%2C%20Thomas%2C%20Y.%2C%20Guo%2C%20Y.%2C%20Stoecklin%2C%20J.%2C%20Neagu%2C%20A.%2C%20Pintard%2C%20L.%2C%20Merlet%2C%20J.%2C%20%26amp%3B%20Ciosk%2C%20R.%20%282018%29.%20PRP-19%2C%20a%20conserved%20pre-mRNA%20processing%20factor%20and%20E3%20ubiquitin%20ligase%2C%20inhibits%20the%20nuclear%20accumulation%20of%20GLP-1%5C%2FNotch%20intracellular%20domain.%20%26lt%3Bi%26gt%3BBiology%20Open%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B7%26lt%3B%5C%2Fi%26gt%3B%287%29%2C%20bio034066.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fbio.034066%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fbio.034066%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22PRP-19%2C%20a%20conserved%20pre-mRNA%20processing%20factor%20and%20E3%20ubiquitin%20ligase%2C%20inhibits%20the%20nuclear%20accumulation%20of%20GLP-1%5C%2FNotch%20intracellular%20domain%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Silvia%22%2C%22lastName%22%3A%22Gutnik%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yann%22%2C%22lastName%22%3A%22Thomas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yanwu%22%2C%22lastName%22%3A%22Guo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Janosch%22%2C%22lastName%22%3A%22Stoecklin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anca%22%2C%22lastName%22%3A%22Neagu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jorge%22%2C%22lastName%22%3A%22Merlet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rafal%22%2C%22lastName%22%3A%22Ciosk%22%7D%5D%2C%22abstractNote%22%3A%22The%20Notch%20signalling%20pathway%20is%20a%20conserved%20and%20widespread%20signalling%20paradigm%2C%20and%20its%20misregulation%20has%20been%20implicated%20in%20numerous%20disorders%2C%20including%20cancer.%20The%20output%20of%20Notch%20signalling%20depends%20on%20the%20nuclear%20accumulation%20of%20the%20Notch%20receptor%20intracellular%20domain%20%28ICD%29.%20Using%20the%20Caenorhabditis%20elegans%20germline%2C%20where%20GLP-1%5C%2FNotch-mediated%20signalling%20is%20essential%20for%20maintaining%20stem%20cells%2C%20we%20monitored%20GLP-1%20in%20vivo%20We%20found%20that%20the%20nuclear%20enrichment%20of%20GLP-1%20ICD%20is%20dynamic%3A%20while%20the%20ICD%20is%20enriched%20in%20germ%20cell%20nuclei%20during%20larval%20development%2C%20it%20is%20depleted%20from%20the%20nuclei%20in%20adult%20germlines.%20We%20found%20that%20this%20pattern%20depends%20on%20the%20ubiquitin%20proteolytic%20system%20and%20the%20splicing%20machinery%20and%2C%20identified%20the%20splicing%20factor%20PRP-19%20as%20a%20candidate%20E3%20ubiquitin%20ligase%20required%20for%20the%20nuclear%20depletion%20of%20GLP-1%20ICD.%22%2C%22date%22%3A%222018-07-16%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1242%5C%2Fbio.034066%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222046-6390%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%227UENQK9Z%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Vigneron%20et%20al.%22%2C%22parsedDate%22%3A%222018-06-04%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVigneron%2C%20S.%2C%20Sundermann%2C%20L.%2C%20Labb%26%23xE9%3B%2C%20J.-C.%2C%20Pintard%2C%20L.%2C%20Radulescu%2C%20O.%2C%20Castro%2C%20A.%2C%20%26amp%3B%20Lorca%2C%20T.%20%282018%29.%20Cyclin%20A-cdk1-Dependent%20Phosphorylation%20of%20Bora%20Is%20the%20Triggering%20Factor%20Promoting%20Mitotic%20Entry.%20%26lt%3Bi%26gt%3BDevelopmental%20Cell%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B45%26lt%3B%5C%2Fi%26gt%3B%285%29%2C%20637-650.e7.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2018.05.005%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2018.05.005%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Cyclin%20A-cdk1-Dependent%20Phosphorylation%20of%20Bora%20Is%20the%20Triggering%20Factor%20Promoting%20Mitotic%20Entry%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Suzanne%22%2C%22lastName%22%3A%22Vigneron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lena%22%2C%22lastName%22%3A%22Sundermann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Claude%22%2C%22lastName%22%3A%22Labb%5Cu00e9%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ovidiu%22%2C%22lastName%22%3A%22Radulescu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thierry%22%2C%22lastName%22%3A%22Lorca%22%7D%5D%2C%22abstractNote%22%3A%22Mitosis%20is%20induced%20by%20the%20activation%20of%20the%20cyclin%20B%5C%2Fcdk1%20feedback%20loop%20that%20creates%20a%20bistable%20state.%20The%20triggering%20factor%20promoting%20active%20cyclin%20B%5C%2Fcdk1%20switch%20has%20been%20assigned%20to%20cyclin%20B%5C%2Fcdk1%20accumulation%20during%20G2.%20However%2C%20this%20complex%20is%20rapidly%20inactivated%20by%20Wee1%5C%2FMyt1-dependent%20phosphorylation%20of%20cdk1%20making%20unlikely%20a%20triggering%20role%20of%20this%20kinase%20in%20mitotic%20commitment.%20Here%20we%20show%20that%20cyclin%20A%5C%2Fcdk1%20kinase%20is%20the%20factor%20triggering%20mitosis.%20Cyclin%20A%5C%2Fcdk1%20phosphorylates%20Bora%20to%20promote%20Aurora%20A-dependent%20Plk1%20phosphorylation%20and%20activation%20and%20mitotic%20entry.%20We%20demonstrate%20that%20Bora%20phosphorylation%20by%20cyclin%20A%5C%2Fcdk1%20is%20both%20necessary%20and%20sufficient%20for%20mitotic%20commitment.%20Finally%2C%20we%20identify%20a%20site%20in%20Bora%20whose%20phosphorylation%20by%20cyclin%20A%5C%2Fcdk1%20is%20required%20for%20mitotic%20entry.%20We%20constructed%20a%20mathematical%20model%20confirming%20the%20essential%20role%20of%20this%20kinase%20in%20mitotic%20commitment.%20Overall%2C%20our%20results%20uncover%20the%20molecular%20mechanism%20by%20which%20cyclin%20A%5C%2Fcdk1%20triggers%20mitotic%20entry.%22%2C%22date%22%3A%222018-06-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.devcel.2018.05.005%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-1551%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%226KAAASYL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Martino%20et%20al.%22%2C%22parsedDate%22%3A%222017-10-23%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BMartino%2C%20L.%2C%20Morchoisne-Bolhy%2C%20S.%2C%20Cheerambathur%2C%20D.%20K.%2C%20Van%20Hove%2C%20L.%2C%20Dumont%2C%20J.%2C%20Joly%2C%20N.%2C%20Desai%2C%20A.%2C%20Doye%2C%20V.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282017%29.%20Channel%20Nucleoporins%20Recruit%20PLK-1%20to%20Nuclear%20Pore%20Complexes%20to%20Direct%20Nuclear%20Envelope%20Breakdown%20in%20C.%26%23xA0%3Belegans.%20%26lt%3Bi%26gt%3BDevelopmental%20Cell%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B43%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20157-171.e7.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.09.019%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.09.019%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Channel%20Nucleoporins%20Recruit%20PLK-1%20to%20Nuclear%20Pore%20Complexes%20to%20Direct%20Nuclear%20Envelope%20Breakdown%20in%20C.%5Cu00a0elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lisa%22%2C%22lastName%22%3A%22Martino%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phanie%22%2C%22lastName%22%3A%22Morchoisne-Bolhy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dhanya%20K.%22%2C%22lastName%22%3A%22Cheerambathur%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Dumont%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arshad%22%2C%22lastName%22%3A%22Desai%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Val%5Cu00e9rie%22%2C%22lastName%22%3A%22Doye%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22In%20animal%20cells%2C%20nuclear%20envelope%20breakdown%20%28NEBD%29%20is%20required%20for%20proper%20chromosome%20segregation.%20Whereas%20mitotic%20kinases%20have%20been%20implicated%20in%20NEBD%2C%20how%20they%20coordinate%20their%20activity%20to%20trigger%20this%20event%20is%20unclear.%20Here%2C%20we%20show%20that%20both%20in%20human%20cells%20and%20Caenorhabditis%20elegans%2C%20the%20Polo-like%20kinase%201%20%28PLK-1%29%20is%20recruited%20to%20the%20nuclear%20pore%20complexes%2C%20just%20prior%20to%20NEBD%2C%20through%20its%20Polo-box%20domain%20%28PBD%29.%20We%20provide%20evidence%20that%20PLK-1%20localization%20to%20the%20nuclear%20envelope%20%28NE%29%20is%20required%20for%20efficient%20NEBD.%20We%20identify%20the%20central%20channel%5Cu00a0nucleoporins%20NPP-1%5C%2FNup58%2C%20NPP-4%5C%2FNup54%2C%20and%20NPP-11%5C%2FNup62%20as%20the%20critical%20factors%20anchoring%20PLK-1%20to%20the%20NE%20in%20C.%5Cu00a0elegans.%20In%20particular%2C%20NPP-1%2C%20NPP-4%2C%20and%20NPP-11%20primed%20at%20multiple%20Polo-docking%20sites%20by%20Cdk1%20and%20PLK-1%20itself%20physically%20interact%20with%20the%20PLK-1%20PBD.%20We%20conclude%20that%20nucleoporins%20play%20an%20unanticipated%20regulatory%20role%20in%20NEBD%2C%20by%20recruiting%20PLK-1%20to%20the%20NE%20thereby%20facilitating%20phosphorylation%20of%20critical%20downstream%20targets.%22%2C%22date%22%3A%222017-10-23%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.devcel.2017.09.019%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-1551%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222023-05-02T12%3A21%3A57Z%22%7D%7D%2C%7B%22key%22%3A%22RWWWZVZ8%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Dickinson%20et%20al.%22%2C%22parsedDate%22%3A%222017-08-21%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BDickinson%2C%20D.%20J.%2C%20Schwager%2C%20F.%2C%20Pintard%2C%20L.%2C%20Gotta%2C%20M.%2C%20%26amp%3B%20Goldstein%2C%20B.%20%282017%29.%20A%20Single-Cell%20Biochemistry%20Approach%20Reveals%20PAR%20Complex%20Dynamics%20during%20Cell%20Polarization.%20%26lt%3Bi%26gt%3BDevelopmental%20Cell%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B42%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20416-434.e11.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.07.024%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.07.024%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20Single-Cell%20Biochemistry%20Approach%20Reveals%20PAR%20Complex%20Dynamics%20during%20Cell%20Polarization%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%20J.%22%2C%22lastName%22%3A%22Dickinson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Francoise%22%2C%22lastName%22%3A%22Schwager%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Monica%22%2C%22lastName%22%3A%22Gotta%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bob%22%2C%22lastName%22%3A%22Goldstein%22%7D%5D%2C%22abstractNote%22%3A%22Regulated%20protein-protein%20interactions%20are%20critical%20for%20cell%20signaling%2C%20differentiation%2C%20and%20development.%20For%20the%20study%20of%20dynamic%20regulation%20of%20protein%20interactions%20in%5Cu00a0vivo%2C%20there%20is%20a%20need%20for%20techniques%20that%20can%20yield%20time-resolved%20information%20and%20probe%20multiple%20protein%20binding%20partners%20simultaneously%2C%20using%20small%20amounts%20of%20starting%20material.%20Here%20we%20describe%20a%20single-cell%20protein%20interaction%20assay.%20Single-cell%20lysates%20are%20generated%20at%20defined%20time%20points%20and%20analyzed%20using%20single-molecule%20pull-down%2C%20yielding%20information%20about%20dynamic%20protein%20complex%20regulation%20in%5Cu00a0vivo.%20We%20established%20the%20utility%20of%20this%20approach%20by%20studying%20PAR%20polarity%20proteins%2C%20which%20mediate%20polarization%20of%20many%20animal%20cell%20types.%20We%20uncovered%20striking%20regulation%20of%20PAR%20complex%20composition%20and%20stoichiometry%20during%20Caenorhabditis%20elegans%20zygote%20polarization%2C%20which%20takes%20place%20in%20less%20than%2020%5Cu00a0min.%20PAR%20complex%20dynamics%20are%20linked%20to%20the%20cell%20cycle%20by%20Polo-like%20kinase%201%20and%20govern%20the%20movement%20of%20PAR%20proteins%20to%20establish%20polarity.%20Our%20results%20demonstrate%20an%20approach%20to%20study%20dynamic%20biochemical%20events%20in%5Cu00a0vivo.%22%2C%22date%22%3A%222017-08-21%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.devcel.2017.07.024%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-1551%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A28%3A03Z%22%7D%7D%2C%7B%22key%22%3A%22CNKI2D58%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Richarme%20et%20al.%22%2C%22parsedDate%22%3A%222017-07-14%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BRicharme%2C%20G.%2C%20Liu%2C%20C.%2C%20Mihoub%2C%20M.%2C%20Abdallah%2C%20J.%2C%20Leger%2C%20T.%2C%20Joly%2C%20N.%2C%20Liebart%2C%20J.-C.%2C%20Jurkunas%2C%20U.%20V.%2C%20Nadal%2C%20M.%2C%20Bouloc%2C%20P.%2C%20Dairou%2C%20J.%2C%20%26amp%3B%20Lamouri%2C%20A.%20%282017%29.%20Guanine%20glycation%20repair%20by%20DJ-1%5C%2FPark7%20and%20its%20bacterial%20homologs.%20%26lt%3Bi%26gt%3BScience%20%28New%20York%2C%20N.Y.%29%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B357%26lt%3B%5C%2Fi%26gt%3B%286347%29%2C%20208%26%23×2013%3B211.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fscience.aag1095%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fscience.aag1095%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Guanine%20glycation%20repair%20by%20DJ-1%5C%2FPark7%20and%20its%20bacterial%20homologs%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gilbert%22%2C%22lastName%22%3A%22Richarme%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cailing%22%2C%22lastName%22%3A%22Liu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mouadh%22%2C%22lastName%22%3A%22Mihoub%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jad%22%2C%22lastName%22%3A%22Abdallah%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thibaut%22%2C%22lastName%22%3A%22Leger%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Claude%22%2C%22lastName%22%3A%22Liebart%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ula%20V.%22%2C%22lastName%22%3A%22Jurkunas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marc%22%2C%22lastName%22%3A%22Nadal%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Philippe%22%2C%22lastName%22%3A%22Bouloc%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Dairou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aazdine%22%2C%22lastName%22%3A%22Lamouri%22%7D%5D%2C%22abstractNote%22%3A%22DNA%20damage%20induced%20by%20reactive%20carbonyls%20%28mainly%20methylglyoxal%20and%20glyoxal%29%2C%20called%20DNA%20glycation%2C%20is%20quantitatively%20as%20important%20as%20oxidative%20damage.%20DNA%20glycation%20is%20associated%20with%20increased%20mutation%20frequency%2C%20DNA%20strand%20breaks%2C%20and%20cytotoxicity.%20However%2C%20in%20contrast%20to%20guanine%20oxidation%20repair%2C%20how%20glycated%20DNA%20is%20repaired%20remains%20undetermined.%20Here%2C%20we%20found%20that%20the%20parkinsonism-associated%20protein%20DJ-1%20and%20its%20bacterial%20homologs%20Hsp31%2C%20YhbO%2C%20and%20YajL%20could%20repair%20methylglyoxal-%20and%20glyoxal-glycated%20nucleotides%20and%20nucleic%20acids.%20DJ-1-depleted%20cells%20displayed%20increased%20levels%20of%20glycated%20DNA%2C%20DNA%20strand%20breaks%2C%20and%20phosphorylated%20p53.%20Deglycase-deficient%20bacterial%20mutants%20displayed%20increased%20levels%20of%20glycated%20DNA%20and%20RNA%20and%20exhibited%20strong%20mutator%20phenotypes.%20Thus%2C%20DJ-1%20and%20its%20prokaryotic%20homologs%20constitute%20a%20major%20nucleotide%20repair%20system%20that%20we%20name%20guanine%20glycation%20repair.%22%2C%22date%22%3A%222017-07-14%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1126%5C%2Fscience.aag1095%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221095-9203%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A28%3A17Z%22%7D%7D%5D%7D
Pillan, A., Ormancey, P., Ben Choug, C., Orlicky, S., Tavernier, N., Van Hove, L., Ossareh-Nazari, B., Joly, N., Sicheri, F., Lorca, T., & Pintard, L. (2026). Molecular basis for the activation of Aurora A and Plk1 kinases during mitotic entry. The EMBO Journal. https://doi.org/10.1038/s44318-025-00679-8
Joly, N., & Pintard, L. (2025). Intramolecular regulation of the MT-severing enzyme Katanin prevents futile ATP hydrolysis. Journal of Cell Biology, 225(2), e202506192. https://doi.org/10.1083/jcb.202506192
Roumbo, L., Ossareh-Nazari, B., Vigneron, S., Stefani, I., Van Hove, L., Legros, V., Chevreux, G., Lacroix, B., Castro, A., Joly, N., Lorca, T., & Pintard, L. (2025). The MAST kinase KIN-4 carries out mitotic entry functions of Greatwall in C. elegans. The EMBO Journal, 1–32. https://doi.org/10.1038/s44318-025-00364-w
El Mossadeq, L., Bellutti, L., Le Borgne, R., Canman, J. C., Pintard, L., Verbavatz, J.-M., Askjaer, P., & Dumont, J. (2024). An interkinetic envelope surrounds chromosomes between meiosis I and II in C. elegans oocytes. Journal of Cell Biology, 224(3), e202403125. https://doi.org/10.1083/jcb.202403125
Strzelecki, P., Joly, N., Hébraud, P., Hoffmann, E., Cech, G. M., Kloska, A., Busi, F., & Grange, W. (2024). Enhanced Golden Gate Assembly: evaluating overhang strength for improved ligation efficiency. Nucleic Acids Research, gkae809. https://doi.org/10.1093/nar/gkae809
Beaumale, E., Van Hove, L., Pintard, L., & Joly, N. (2024). Microtubule-binding domains in Katanin p80 subunit are essential for severing activity in C. elegans. Journal of Cell Biology, 223(4), e202308023. https://doi.org/10.1083/jcb.202308023
Velez-Aguilera, G., Ossareh-Nazari, B., & Pintard, L. (2024). Dissecting the Multiple Functions of the Polo-Like Kinase 1 in the C. elegans Zygote. In A. Castro & B. Lacroix (Eds.), Cell Cycle Control: Methods and Protocols (pp. 63–88). Springer US. https://doi.org/10.1007/978-1-0716-3557-5_4
Nkombo Nkoula, S., Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Ayuso, C., Legros, V., Chevreux, G., Thomas, L., Seydoux, G., Askjaer, P., & Pintard, L. (2023). Mechanisms of nuclear pore complex disassembly by the mitotic Polo-like kinase 1 (PLK-1) in C. elegans embryos. Science Advances, 9(29), eadf7826. https://doi.org/10.1126/sciadv.adf7826
Kouranti, I., Abdel Khalek, W., Mazurkiewicz, S., Loisel-Ferreira, I., Gautreau, A. M., Pintard, L., Jeunemaitre, X., & Clauser, E. (2022). Cullin 3 Exon 9 Deletion in Familial Hyperkalemic Hypertension Impairs Cullin3-Ring-E3 Ligase (CRL3) Dynamic Regulation and Cycling. International Journal of Molecular Sciences, 23(9), 5151. https://doi.org/10.3390/ijms23095151
Pillan, A., Tavernier, N., & Pintard, L. (2022). [The kiss of life: Aurora A embraces the phosphate of its cofactor Bora to trigger mitotic entry]. Medecine Sciences: M/S, 38(4), 345–347. https://doi.org/10.1051/medsci/2022033
Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Joly, N., & Pintard, L. (2022). Cortical microtubule pulling forces contribute to the union of the parental genomes in the Caenorhabditis elegans zygote. eLife, 11, e75382. https://doi.org/10.7554/eLife.75382
Tavernier, N., Sicheri, F., & Pintard, L. (2021). Aurora A kinase activation: Different means to different ends. The Journal of Cell Biology, 220(9), e202106128. https://doi.org/10.1083/jcb.202106128
Knox, J., Joly, N., Linossi, E. M., Carmona-Negrón, J. A., Jura, N., Pintard, L., Zuercher, W., & Roy, P. J. (2021). A survey of the kinome pharmacopeia reveals multiple scaffolds and targets for the development of novel anthelmintics. Scientific Reports, 11(1), 9161. https://doi.org/10.1038/s41598-021-88150-6
Tavernier, N., Thomas, Y., Vigneron, S., Maisonneuve, P., Orlicky, S., Mader, P., Regmi, S. G., Van Hove, L., Levinson, N. M., Gasmi-Seabrook, G., Joly, N., Poteau, M., Velez-Aguilera, G., Gavet, O., Castro, A., Dasso, M., Lorca, T., Sicheri, F., & Pintard, L. (2021). Bora phosphorylation substitutes in trans for T-loop phosphorylation in Aurora A to promote mitotic entry. Nature Communications, 12(1), 1899. https://doi.org/10.1038/s41467-021-21922-w
Velez-Aguilera, G., Nkombo Nkoula, S., Ossareh-Nazari, B., Link, J., Paouneskou, D., Van Hove, L., Joly, N., Tavernier, N., Verbavatz, J.-M., Jantsch, V., & Pintard, L. (2020). PLK-1 promotes the merger of the parental genome into a single nucleus by triggering lamina disassembly. eLife, 9, e59510. https://doi.org/10.7554/eLife.59510
Joly, N., Beaumale, E., Van Hove, L., Martino, L., & Pintard, L. (2020). Phosphorylation of the microtubule-severing AAA+ enzyme Katanin regulates C. elegans embryo development. The Journal of Cell Biology, 219(6), e201912037. https://doi.org/10.1083/jcb.201912037
Pintard, L., & Bowerman, B. (2019). Mitotic Cell Division in Caenorhabditis elegans. Genetics, 211(1), 35–73. https://doi.org/10.1534/genetics.118.301367
Pintard, L., & Archambault, V. (2018). A unified view of spatio-temporal control of mitotic entry: Polo kinase as the key. Open Biology, 8(8), 180114. https://doi.org/10.1098/rsob.180114
Gutnik, S., Thomas, Y., Guo, Y., Stoecklin, J., Neagu, A., Pintard, L., Merlet, J., & Ciosk, R. (2018). PRP-19, a conserved pre-mRNA processing factor and E3 ubiquitin ligase, inhibits the nuclear accumulation of GLP-1/Notch intracellular domain. Biology Open, 7(7), bio034066. https://doi.org/10.1242/bio.034066
Vigneron, S., Sundermann, L., Labbé, J.-C., Pintard, L., Radulescu, O., Castro, A., & Lorca, T. (2018). Cyclin A-cdk1-Dependent Phosphorylation of Bora Is the Triggering Factor Promoting Mitotic Entry. Developmental Cell, 45(5), 637-650.e7. https://doi.org/10.1016/j.devcel.2018.05.005
Martino, L., Morchoisne-Bolhy, S., Cheerambathur, D. K., Van Hove, L., Dumont, J., Joly, N., Desai, A., Doye, V., & Pintard, L. (2017). Channel Nucleoporins Recruit PLK-1 to Nuclear Pore Complexes to Direct Nuclear Envelope Breakdown in C. elegans. Developmental Cell, 43(2), 157-171.e7. https://doi.org/10.1016/j.devcel.2017.09.019
Dickinson, D. J., Schwager, F., Pintard, L., Gotta, M., & Goldstein, B. (2017). A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization. Developmental Cell, 42(4), 416-434.e11. https://doi.org/10.1016/j.devcel.2017.07.024
Richarme, G., Liu, C., Mihoub, M., Abdallah, J., Leger, T., Joly, N., Liebart, J.-C., Jurkunas, U. V., Nadal, M., Bouloc, P., Dairou, J., & Lamouri, A. (2017). Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science (New York, N.Y.), 357(6347), 208–211. https://doi.org/10.1126/science.aag1095
- Frank SICHERI (University of Toronto, Canada)
- Thierry LORCA (CRBM, Montpellier, France)
- Anna CASTRO (CRBM, Montpellier, France)
- Olivier GAVET (Institut Gustave Roussy, Paris Villejuif)
- Mary DASSO (NIH, Bethesda USA)
- Peter ASKJAER (CABD, Sevilla, Spain)
- Verena JANTSCH (Max Perutz Labs Vienna, Austria)
- Antoine JEGOU (Institut Jacques Monod, Paris, France)
- Guillaume ROMET-LEMONNE (Institut Jacques Monod, Paris, France)
- Denis CHRETIEN (IGDR, Rennes, France)
- Julien DUMONT (Institut Jacques Monod, Paris, France)
- Valérie DOYE (Institut Jacques Monod, Paris, France)
- Bruce BOWERMAN (IMB, University of Oregon, USA)
- Arshad DESAI (USCD, San Diego, USA)
- Monica GOTTA (University of Geneva, Switzerland)
- ANR AMBRE
- ANR REPLIGREAT
- ARC
- Idex « AAP Dynamique » Université de Paris
- Equipe Labellisée Ligue contre le Cancer
- Labex « WHO AM I »
30/01/2026 : Scientific news published by CNRS Biology : Bora, le chef d’orchestre moléculaire qui déclenche l’entrée en mitose
26/11/2025 : Scientific news published by CNRS Biology: Un mécanisme d’auto-régulation empêche la Katanine de gaspiller l’énergie cellulaire
25/10/2021 : Nicolas JOLY (Institut Jacques Monod), winner of the Maurice Nicloux Prize 2021!
Master and PhD students
Our laboratory offers a wide range of interesting and challenging research projects for motivated master or PhD candidates aimed at understanding the mechanisms of cell cycle control during animal development.
We employ a unique combination of genetics, biochemistry, cell biology, live imaging, quantitative proteomics, and functional genomics approaches.
PostDoc candidates
We are looking for highly motivated and team-oriented scientists with a strong background in biochemistry, genetics and cell biology.
Candidates are welcome to apply with CV, publication list, motivation letter, and names of 2 referees to:
Lionel PINTARD – Université Paris cité, CNRS, Institut Jacques Monod, 15 rue Hélène Brion – 75013 PARIS cedex 13 – France


