RNA biogenesis and genome homeostasis
Benoit PALANCADE
Guaranteeing genome integrity while ensuring gene expression is a permanent challenge in living cells: while the regulated production of messenger RNAs is critical for the establishment of gene expression patterns, it can compete with the maintenance and the duplication of chromosomes. In this context, our team explores the mechanisms that keep in check the different stages of mRNA metabolism, from their synthesis in the nucleus to their export and translation in the cytoplasm.
Keywords: messenger RNA ; gene expression ; transcription ; genetic stability ; DNA:RNA hybrids and R-loops ; localized translation ; sumoylation ; nuclear pore complexes.
+33 (0)1 57 27 80 39 Contact @palancadelab.bsky.social
Guaranteeing genome integrity while ensuring gene expression is a permanent challenge in living cells: while the regulated production of messenger RNAs is critical for the establishment of gene expression patterns, it can compete with the maintenance and the duplication of chromosomes. In this context, our team explores the mechanisms that keep in check the different stages of mRNA metabolism, from their synthesis in the nucleus to their export and translation in the cytoplasm. We are specially willing to understand:
- how mRNA biogenesis is synchronized with other DNA-dependent molecular transactions, by counteracting the formation of genotoxic DNA:mRNA hybrids (or R-loops);
- how these processes are spatially organized, in particular with respect to nuclear pores, the macromolecular complexes ensuring the transit of mRNAs out of the nucleus.
To address these questions, the integrative approach adopted by our team combines systematic and mechanistic approaches in a unicellular model eukaryote, the yeast S. cerevisiae .
The synchronization of the different activities targeting the genetic material in the nucleus is essential to maintain genome homeostasis. In this context, mRNA metabolism has recently emerged as an essential player, not only in the gene expression process, but also in the maintenance of genetic stability (Fig. 1). On the one hand, the combinatorial association of RNA-binding proteins (RBPs) with mRNAs controls their synthesis, processing, transport and degradation, ultimately defining their cellular fate and translation into proteins. In addition, mRNA biogenesis counteracts the formation of “R-loops”: these structures, which encompass a DNA:mRNA hybrid together with a displaced single-stranded DNA moiety, can increase the sensitivity of the genetic material to genotoxic agents, and interfere with its duplication, thus representing a source of genetic instability. To precisely control mRNA biogenesis, the activities of several multiprotein complexes are coupled and regulated from the synthesis of the transcripts to their export out of the nucleus and their translation. Conversely, uncoupling the different steps of the mRNA biogenesis process has deleterious effects for both gene expression and genome stability, as observed in multiple physiological, mutant or pathological situations (e.g. cancer).
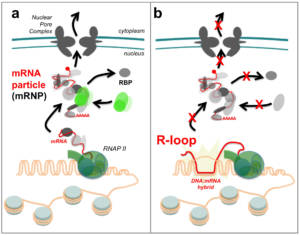
Figure 1 – The biogenesis of mRNA particles (mRNPs) and its impact on gene expression and genome stability. a, Synchronization of the different stages of mRNA biogenesis : the recruitment of RNA-binding proteins (RBP) specifies the cellular fate of the transcripts, and counteracts the formation of R-loops. b, Uncoupling the different stages of mRNA biogenesis : defects in mRNA processing or export lead to the formation of genotoxic R-loops. RNAP II, RNA polymerase II. Copyright CNRS / IJM / B. PALANCADE.
The team’s research activities aim to identify the cis- and trans-acting regulatory mechanisms targeting mRNA metabolism, and to understand their impact on genome expression and integrity. We are particularly interested in (i) the influence of mRNA biogenesis on the formation of R-loops and its consequences on the maintenance of genome structure and stability (see below, axis 1), and in (ii) the spatial organization of these processes, in particular with respect to nuclear pores, the macromolecular complexes involved in the transport of mRNAs to the cytoplasm (see below, axis 2). To this end, systematic approaches – genomics and proteomics – are combined with mechanistic studies in vitro and in vivo, mainly in the budding yeast S. cerevisiae, a model system for understanding these conserved processes (Fig. 2).
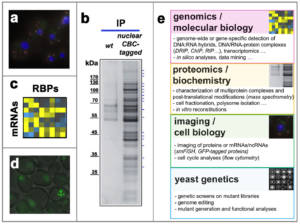
Figure 2 – RNA biogenesis and genome homeostasis: tools and approaches. a, The subcellular localization of mRNAs – here, those encoding nuclear pore subunits – is analyzed by smFISH (single molecule fluorescence in situ hybridization) in yeast cells. b, The composition of mRNA particles is analyzed by mass spectrometry following their affinity purification using a tagged version of the cap–binding complex (CBC ; silver staining after denaturing electrophoresis). c, The transcriptome-wide association of RBPs with mRNAs can be compiled following interaction data mining. d, The formation of repair foci labeled by a fluorescent version of Rad52 is detected by live imaging on yeast cells inactivated for an RNA biogenesis factor (THO complex), revealing the genetic instability typical of this mutant. e, Overview of the approaches used in the lab. Copyright CNRS / IJM / B. PALANCADE.
Axis 1 – mRNA biogenesis and the maintenance of genetic integrity: the R-loops under control
While the formation of R-loops has recently been shown to be widespread in the tree of life, and associated with multiple genomic processes, our understanding of the cellular mechanisms that modulate their accumulation remains limited. To identify the factors that control the formation of these structures in cis and in trans, we have combined genomic and genetic approaches in yeast, thus highlighting novel aspects of their regulation.
1.1. A new role for introns: protecting eukaryotic genomes from R-loop-associated genetic instability
The central role of intron splicing in the control of genome expression has led us to assess its impact on mRNA biogenesis and genetic stability. For this, we performed a screen based on a reporter system to identify the factors acting specifically in the biogenesis and the export of mRNAs from intron-containing genes (Bonnet et al., 2015 ; see also Bonnet & Palancade, 2015). This study revealed that the presence of introns attenuated the deleterious effect of mutations associated with the accumulation of R-loops, suggesting that these sequences could counteract their genotoxicity. To expand these observations, we explored genome-wide maps of R-loops and DNA damage distribution in S. cerevisiae and human cells, and showed that these genetic lesions preferentially accumulate on intronless genes (collaboration with Sergio de ALMEIDA, University of Lisbon, Portugal). By directly modifying the intron content of model genes through genome editing approaches in yeast, we then established that the removal of endogenous introns leads to the appearance of R-loops, while the insertion of introns into naturally intronless genes attenuates the formation of these structures and the associated DNA damages. Insertion in place of an intron of different sequences capable of splicing and/or associating with proteins ultimately revealed that it is the binding of proteins to RNA, and not the splicing process per se, which sterically opposes the formation of DNA:RNA hybrids. This novel function of introns in protecting against genotoxic structures could contribute to explain their conservation during evolution (Fig. 3; Bonnet et al., 2017 ; Palancade, 2018).
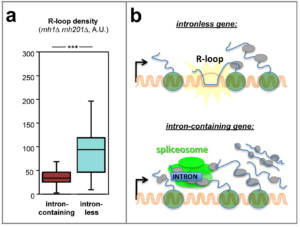
Figure 3 – Introns protect genomes from the accumulation of R-loops. a, Decreased levels of DNA:RNA hybrids on S. cerevisiae intron-containing genes in genomic studies (analysis performed on cells inactivated for RNases H, which naturally restrict hybrid accumulation ). b, Model: on intron-containing genes, the recruitment of the spliceosome physically counteracts the formation of DNA:RNA hybrids. According to Bonnet et al, 2017. Copyright CNRS / IJM / B. PALANCADE.
1.2. Towards a systematic depiction of the pathways controlling R-loop formation and resolution
Despite the growing interest of the scientific community for the mechanisms of R-loop formation and genotoxicity, none systematic approach has been used so far to identify the cellular factors controlling the metabolism of these structures. In collaboration with the team of Rodney ROTHSTEIN (Columbia University Medical Center, NY, USA), we have designed a genetic screen to identify the proteins preventing the formation of R-loops or ensuring their resolution, thus limiting the associated genetic instability. Surprisingly, several of the identified factors are associated with other genomic processes, confirming the close relationships between RNA and DNA metabolism, as already attested by the existence of cellular machineries that do not strictly discriminate between these different substrates (Palancade & Rothstein, 2021). The mechanistic characterization of the factors thereby identified should ultimately allow to understand how mRNA biogenesis is synchronized with DNA-related transactions, in particular replication and repair.
Axis 2 – The spatial organization of mRNA metabolism: a central role for nuclear pores
2.1. Nuclear pores and mRNA biogenesis: a matter of SUMOylation
When mRNA particles (mRNPs) enter the export pathway on the nucleoplasmic face of nuclear pore complexes (NPCs), they are found in close proximity with Ulp1SENP2, a SUMO-deconjugating enzyme that regulates post-translational modifications by this small polypeptide (Palancade & Doye, 2008). To determine whether SUMOylation thus modulates mRNA biogenesis or transport, we have combined data mining (Rouviere et al, 2013; Rouviere et al, 2018), proteomics (Bretes et al, 2014) and transcriptomics (Bonnet et al, 2021). Following a proteomic screen designed to characterize the composition of mRNPs at different stages of their biogenesis (collaboration with Marlene OEFFINGER, IRCM, Montreal, Canada, and with the ProteoSeine Proteomics platform at IJM), we have notably shown that SUMO controls the recruitment of a limited number of RNA-binding proteins, including the conserved factors THO and Hek2hnRNP-K. In the case of the THO complex, a key mRNA biogenesis factor, this SUMO-dependent recruitment counteracts the degradation of certain stress-induced mRNAs, ensuring survival in such situations (Fig. 4 ; Bretes et al., 2014). On the other hand, the regulated association of Hek2 with its target mRNAs allows their translational repression (collaboration with Bertrand COSSON, University of Paris). Strikingly, the mRNAs encoding NPC subunits are themselves the main targets of this regulation, revealing a feedback loop in which Ulp1 and Hek2 are respectively the sensor and the effector of a mechanism maintaining the integrity of nuclear pores (Fig. 5 ; Rouviere et al, 2018). Beyond these post-transcriptional regulations, our transcriptomic studies also revealed that NPC-dependent (de)SUMOylation mechanisms could control the synthesis of other classes of mRNA (Bonnet et al, 2015), in particular by repressing the transcription of transposable elements (collaboration with Pascale LESAGE, Hôpital St-Louis, Paris ; Bonnet et al., 2021).
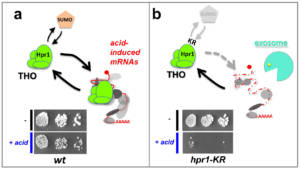
Figure 4 – SUMOylation regulates stress-induced mRNA biogenesis and stability. a, In wild type cells, the SUMOylation of the THO complex allows its association with acidic stress-induced mRNAs. The expression of such mRNAs thus allows survival under these specific stress conditions (bottom panel). b , In mutant cells impaired for THO complex SUMOylation on its Hpr1 subunit (hpr1-KR ), acidic stress-induced mRNAs exhibit decreased association with the THO complex and exosome-dependent degradation, resulting in a loss of viability under the stress conditions (bottom panel). According to Bretes et al. 2014. Copyright CNRS / IJM / B. PALANCADE.
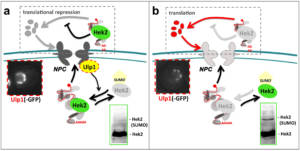
Figure 5 – A regulatory loop controlling the integrity of a multiprotein complex. a, When nuclear pore complexes (NPCs) are properly assembled, elevated levels of the NPC-associated SUMO-deconjugating enzyme Ulp1 causes de-SUMOylation of the Hek2 protein and its binding to mRNAs encoding subunits of NPCs, repressing their translation. b, In case of defective NPCs, decreased Ulp1 levels leads to enhanced Hek2 SUMOylation, reduced levels of Hek2 association with its target mRNAs and subsequent translation of nuclear pore subunits. According to Rouviere et al, 2018. Copyright CNRS / IJM / B. PALANCADE.
2.2. mRNA translation at nuclear pores
The identification of translational regulations collectively targeting the mRNAs encoding the subunits of a protein complex prompted us to tackle the coupling between polypeptide synthesis and assembly. By systematically analyzing the co-translational interactions occurring between NPC subunits, we have thus established that certain NPC building blocks assemble in an ordered manner, during the translation of their constituents, in the cytoplasm. On the other hand, we found that certain mRNAs encoding nucleoplasmically-oriented NPC subunits or other nuclear proteins are translated at the vicinity of nuclear pores (collaboration with Martine COLLART, University of Geneva, Switzerland, and Frédéric DEVAUX, Sorbonne Université, Paris). Localized translation requires the recognition of nuclear targeting signals in nascent polypeptides by transport factors themselves associated with NPCs, the karyopherins, and would prevent the deleterious accumulation of toxic or aggregation-prone proteins in the cytoplasm (Lautier et al, 2021). In this view, these localized translation events are reminiscent of those targeting signal peptide-containing proteins to the endoplasmic reticulum, although the associated machineries have distinct evolutionary origins (Fig. 6).
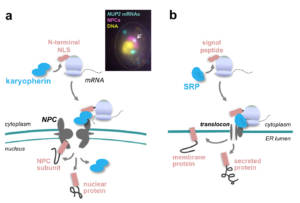
Figure 6 – mRNA translation at nuclear pores. a, Schematic representation of the mechanism targeting mRNAs for translation at NPCs, and typical in situ hybridization image showing the localization of the NUP2 mRNA. b, Comparison with the targeting mechanism of mRNAs encoding membrane or secreted proteins at the endoplasmic reticulum (ER). In the latter case, localized translation requires the SRP (signal recognition particle)-dependent recognition of the signal peptide in the nascent protein. Copyright CNRS / IJM / B. PALANCADE.
Other contributions in the fields of mRNA biogenesis and genome homeostasis
As part of the work described above, we have implemented different approaches to study mRNA metabolism and its impact on genome stability, which have also been used in the frame of collaborations. Hence, we have participated in various studies by functionally characterizing ribonucleoprotein complexes or SUMO post-translational modifications (Infantino et al., 2019; Talhaoui et al., 2018; Delaveau et al, 2016), by monitoring RNA localization in situ (Andric et al., 2021; Kramarz et al, 2020; Hazra et al, 2020) or by measuring genetic instability with dedicated reporters (Babour et al., 2016 ; Fritzen et al., al., 2016).
Members
 Anna BABOUR, Researcher, PALANCADE LAB+33 (0)1 57 27 80 39, room 354B
Anna BABOUR, Researcher, PALANCADE LAB+33 (0)1 57 27 80 39, room 354B Matteo BURELLI, Intern, PALANCADE LAB
Matteo BURELLI, Intern, PALANCADE LAB Karine CASIER, Biology engineer, PALANCADE LAB+33 (0)1 57 27 80 39, room 354B
Karine CASIER, Biology engineer, PALANCADE LAB+33 (0)1 57 27 80 39, room 354B Coralie GONCALVES PEREIRA PERRUCA, Assistant biology engineer, PALANCADE LAB+33 (0)1 57 27 80 38, room 354B
Coralie GONCALVES PEREIRA PERRUCA, Assistant biology engineer, PALANCADE LAB+33 (0)1 57 27 80 38, room 354B Mohamed MLIH, Biology engineer, PALANCADE LAB+33 (0)1 57 27 80 38, room 354B
Mohamed MLIH, Biology engineer, PALANCADE LAB+33 (0)1 57 27 80 38, room 354B Emma THOMAS, PhD student, PALANCADE LAB+33 (0)1 57 27 80 38, room 354B
Emma THOMAS, PhD student, PALANCADE LAB+33 (0)1 57 27 80 38, room 354B Myriam ZHENG, PhD student, PALANCADE LAB+33 (0)1 57 27 80 39, room 354B
Myriam ZHENG, PhD student, PALANCADE LAB+33 (0)1 57 27 80 39, room 354B
To contact a member of the team by e-mail: name.surname@ijm.fr
- Mangione RM, Pierce S, Zheng M, Martin RM, Goncalves C, Kumar A, Scaglione S, de Sousa Morgado C, Penzo A, Lancrey A, Reid RJD, Lautier O, Gaillard PH, Stirling PC, de Almeida SF, Rothstein R, Palancade B. DNA lesions can frequently precede DNA:RNA hybrid accumulation. Nat Commun. 2025 Mar 10;16(1):2401. doi: 10.1038/s41467-025-57588-x. PMID: 40064914.
- Penzo, A., Dubarry, M., Brocas, C., Zheng, M., Mangione, R. M., Rougemaille, M., Goncalves, C., Lautier, O., Libri, D., Simon, M.-N., Géli, V., Dubrana, K., & Palancade, B. (2023). A R-loop sensing pathway mediates the relocation of transcribed genes to nuclear pore complexes. Nature Communications, 14(1), 5606.
- Aiello, U., Challal, D., Wentzinger, G., Lengronne, A., Appanah, R., Pasero, P., Palancade, B., & Libri, D. (2022). Sen1 is a key regulator of transcription-driven conflicts. Molecular Cell, 82(16), 2952-2966.e6.
- Bonnet A, Chaput C, Palmic N, Palancade B* & Lesage P* (2021) A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription. PLoS Genet. 17(11):e1009889
- Lautier O, Penzo A, Rouvière JO, Chevreux G, Collet L, Loïodice I, Taddei A, Devaux F, Collart MA, Palancade B. (2021) Co-translational assembly and localized translation of nucleoporins in nuclear pore complex biogenesis. Mol Cell. 2021 Apr 4:S1097-2765(21)00225-2.
- Cover caption
- See news from CNRS (INSB) and Université Paris Cité.
- Rouviere JO, Bulfoni M, Tuck A, Cosson B, Devaux F & Palancade B. (2018) A SUMO-dependent feedback loop senses and controls the biogenesis of nuclear pore subunits. Nature communications 9(1):1665
- Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Géli V, de Almeida SF & Palancade B. (2017) Introns protect eukaryotic genomes from transcription-associated genetic instability Mol Cell 67(4):608-621.e6
- See the commentary in Science and news from CNRS (INSB)
- This article was selected “article de l’année 2017” by the SFBBM (French Society of Biochemistry and Molecular Biology)
- Bonnet A, Bretes H & Palancade B.(2015) Nuclear pores affect distinct stages of intron-containing gene expression Nucleic Acids Research 43(8):4249-61
- Bretes H, Rouviere JO, Léger T, Oeffinger M, Devaux F, Doye V and Palancade B. (2014) Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs Nucleic Acids Research 42(8):5043-58
Publications
- Emma Thomas (since October 2024), Université Paris Cité.
- Myriam Zheng (since October 2022), Université Paris Cité. Genotoxic versus regulatory R-loops: characterization and mechanisms.
- Raphaël Mangione (2020-2024), Université Paris Cité. Functional characterization of novel factors controlling R-loops dynamics, a source of transcription-associated genetic instability.

- Arianna Penzo (2018-2022), Université Paris Cité. Exploring novel mechanisms controlling R-loop-dependent genetic instability
- Jérome Rouvière (2012-2016), Université Paris XI. Study of the role of SUMOylation in the metabolism of mRNA ribonucleoparticles (mRNPs)
- Hugo Bretes Rodrigues (2008-2012), Université Paris XI. Study of the regulation of messenger RNA metabolism in the yeast Saccharomyces cerevisiae.
Collaborations en cours
- Sergio de ALMEIDA, Instituto de Medicina Molecular, University of Lisboa, Portugal
- Frédéric DEVAUX & Mathilde GARCIA, Sorbonne Université, Paris, France
- Karine DUBRANA, Commissariat à l’Energie Atomique, Fontenay-aux-Roses, France
- Gilles FISCHER, Sorbonne Université, Paris, France
- Pierre-Henri GAILLARD, Centre de Recherche en Cancérologie de Marseille, France
- Vincent GELI, Centre de Recherche en Cancérologie de Marseille, France
- Jean-René HUYNH & Anahi MOLLA-HERMAN, Collège de France, Paris, France
- Pascale LESAGE, Hôpital St-Louis, Paris, France
- Domenico LIBRI, Institut Jacques Monod, Paris, France
- Rodney ROTHSTEIN, Columbia University Medical Center, New York, USA
- Mathieu ROUGEMAILLE, I2BC, Gif s/Yvette, France
Collaborations passées
- Martine COLLART, Faculté de Médecine, Université de Genève, Switzerland
- Bertrand COSSON, Epigenetics, Université de Paris, France
- Guilhem JANBON, Institut Pasteur, Paris, France
- Gerard MAZON, Institut Gustave Roussy, Villejuif, France
- Marlene OEFFINGER, IRCM, Montréal, Canada
- Sébastien STORCK, Hôpital Necker, Paris, France
- Françoise STUTZ, Université de Genève (Sciences III), Switzerland
Research in the team is supported by the following public institutions, funding agencies and charities:

- Lautier O, Penzo A, Rouvière JO, Chevreux G, Collet L, Loïodice I, Taddei A, Devaux F, Collart MA, Palancade B. (2021) Co-translational assembly and localized translation of nucleoporins in nuclear pore complex biogenesis. Mol Cell. 2021 Apr 4:S1097-2765(21)00225-2.
- Cover caption
- See news from CNRS (INSB) and Université Paris Cité.
- Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Géli V, de Almeida SF & Palancade B. (2017) Introns protect eukaryotic genomes from transcription-associated genetic instability Mol Cell 67(4):608-621.e6
- See the commentary in Science and news from CNRS (INSB):
- This article was selected “article of the year 2017” par la SFBBM (French Society of Biochemistry and Molecular Biology)


