Cellular Spatial Organization
NICOLAS MINC
Our team aims to understand the basis of cellular organization, by addressing questions including:
- How do cells determine their functional morphologies?
- How might they probe their own size and shape?
- How do these geometrical properties contribute to regulate cell division, growth and tissue architecture?
Keywords: Cell division, Cytoskeleton, Morphogenesis, Embryo development.
+33 (0)1 57 27 80 52 Contact @minclab.bsky.social http://www.minclab.fr/
The spatial organization of cells is key to regulate many fundamental biological processes, including, cell migration, division, embryo development and tissue repair. Defects in cellular organization are often associated with the emergence of developmental defects, and severe diseases like cancer.
Our team aims to understand the basis of cellular organization, by addressing questions including: How do cells determine their functional morphologies? How might they probe their own size and shape? How do these geometrical properties contribute to regulate cell division, growth and tissue architecture?
To reach these goals we combine expertise from different fields including chemistry, biology and physics. We integrate and develop state of the art quantitative approaches, such as micro-fabrication, mathematical modeling and image analysis tools, with more traditional genetics and biochemistry approaches. We aim to establish quantitative rules that regulate morphogenesis in unicellular and multicellular organisms.
Cells come with stereotypical shapes and sizes which are often associated with their function. Neurons grow in highly elongated morphologies to build neuronal networks, while red blood cells have discoid-like deformable morphologies to squeeze within tiny blood vessels. Questions that fascinate our team are to understand how cells define their particular shape, and how information in cell geometry may be conveyed into cell function or in organizing the cell interior. To address these fundamental questions, we integrate original methods at the frontier between molecular biology, genetics and physics and mathematics. One of our ultimate goal is to capture biological behavior with mathematical models. The current main research area of our team include:
1- Division plane positioning and early embryo development
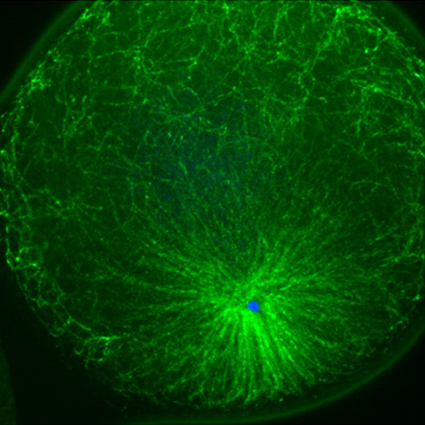
The proper positioning of the site of cell division is crucial not only for the survival of all cells, but also for the development of multi-cellular tissues. The lab is fascinated by the process of early embryogenesis during which the egg becomes fertilized and follows a stereotypical choreography of successive division, called cleavage patterns. Our goal is to understand, predict and establish quantitative laws that explain the patterning of cell divisions in early embryos. To this end, we combine mathematical modelling, imaging and biophysics approaches (Figure 1). Our model experimental system is the sea urchin embryo, which integrates many advantages for the study of early development. To extend our findings to other multicellular embryos and tissues, we also have collaboration with other labs, working on flies, frogs, fishes and ascidians, as well as organoids.
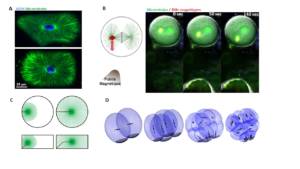
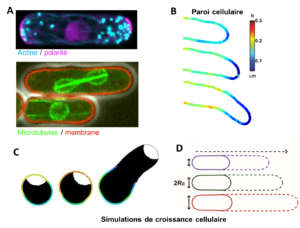
Figure 1: A. In sea urchin zygotes, like in many cells, microtubule asters allow to probe cell shape to orient the nucleus and the subsequent division axis with respect to cell shape (Minc et al. 2011). B. Use of magnetic tweezers to apply forces on mitotic spindles in intact cells, and to control division plane orientation (Xie et al. 2021). C. Numerical simulation of the forces exerted by microtubules to center the nucleus at fertilization (Tanimoto et al. 2016). D. Simulation of the successive division geometries that mark the cleavage stage of early sea urchin embryos (Pierre et al. 2016).
Legend : Developping sea urchin embryos where DNA is labelled with a fluorescent dye.
2- Cell shape and polarity
The morphology of single cells as well as cells embedded in tissues underlie many of their function. We seek to address how cells develop their particular shape and internal organization. For instance, many cells only grow at a given location, a process called polarized growth, which involves specific biochemical reactions needed to cluster a local domain of growth activity (Vidéo 2). To study these aspects we use a unicellular fungi called fission yeast. These cells have rod-shapes and are genetically tractable, with many mutants defective in morphogenesis. These cells are encased in a stiff cell wall and are turgid, which provides them with particular mechanical properties (Figure 2). One of our ultimate goal is to link biochemical and biomechanical signals which serve to shape cells.
Figure 2: A. Fluorescent images of yeast cells expressing fluorescent proteins to visualize the cytoskeleton, polarity, growth and cell division (Bonazzi et al. 2015). B. Super-resolution images of cell wall thickness dynamics in a growin hyphae of the fungus aspergillus nidulans (Chevallier et al). C. Numerical simulation of a yeast spore and its cell wall (colors) and poalrity domain (white). D. Simulation of growth speed f yeast cells.
Members
 Maria Isabel ARJONA HIDALGO, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B
Maria Isabel ARJONA HIDALGO, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B Rossana BETTONI, Postdoctoral researcher, MINC LAB
Rossana BETTONI, Postdoctoral researcher, MINC LAB Serge DMITRIEFF, Researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B
Serge DMITRIEFF, Researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B Amir KHOSRAVANIZADEH, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B
Amir KHOSRAVANIZADEH, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B Charlotte MALLART, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B
Charlotte MALLART, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B Equipe MINC, MINC LAB
Equipe MINC, MINC LAB Ahlam MOULOUDI, Intern, MINC LAB
Ahlam MOULOUDI, Intern, MINC LAB Celia Maria MUNICIO DIAZ, Biology engineer, MINC LAB+33 (0)1 57 27 80 52, room 316B
Celia Maria MUNICIO DIAZ, Biology engineer, MINC LAB+33 (0)1 57 27 80 52, room 316B Tin-Wai NG, Postdoctoral researcher, MINC LAB
Tin-Wai NG, Postdoctoral researcher, MINC LAB Aude NOMMICK, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B
Aude NOMMICK, Postdoctoral researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B Yannis REIGNIER, PhD student, MINC LAB+33 (0)1 57 27 80 52, room 316B
Yannis REIGNIER, PhD student, MINC LAB+33 (0)1 57 27 80 52, room 316B Antoine REMOND, Biology engineer, MINC LAB
Antoine REMOND, Biology engineer, MINC LAB Jérémy SALLE, Researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B
Jérémy SALLE, Researcher, MINC LAB+33 (0)1 57 27 80 52, room 316B Joseph VERMEIL, Postdoctoral researcher, MINC LAB
Joseph VERMEIL, Postdoctoral researcher, MINC LAB Jiawei XU, PhD student, MINC LAB+33 (0)157278052, room 316B
Jiawei XU, PhD student, MINC LAB+33 (0)157278052, room 316B Zhengdunhuang XU, Intern, MINC LAB+33 (0)157278013
Zhengdunhuang XU, Intern, MINC LAB+33 (0)157278013
To contact a member of the team by e-mail: name.surname@ijm.fr
- Xie J, Najafi J, Le Borgne R, Verbavatz J-M, Durieu C, Sallé J, and Minc N (2022) “Contribution of cytoplasm viscoelastic properties to mitotic spindle positioning” , Proc Natl Acad Sci U S A, 119 (8) e2115593119.
- Neeli-Venkata R, Municio Diaz C, Celador R, Sanchez Y, Minc N (2021) “Detection of surface forces by the cell-wall mechanosensor Wsc1 in yeast” , Developmental Cell 56, 1–15
- Palenzuela H, Lacroix B, Sallé J, Minami K, Shima T, Jegou A, Romet-Lemonne#G and Minc# N (2020) “In Vitro Reconstitution of Dynein Force Exertion in a Bulk Viscous Medium” , Current Biology, 30, 1–7
- Davi V, Chevalier L, Guo H, Tanimoto H, Barrett K, Couturier E, Boudaoud#A, and Minc# N, (2019) “Systematic mapping of cell wall mechanics in the regulation of cell morphogenesis” , Proc. Acad. Sci. USA, 116(28):13833-13838
- Sallé J, Xie J, Ershov D, Lacassin M, Dmitrieff S and Minc N, (2019) “Asymmetric division through a reduction of microtubule centering forces” J Cell Biol. 218(3):771-782 .
- Tanimoto H, Sallé J, Dodin L and Minc N (2018) “Physical forces determining the persistency and centring precision of microtubule asters” Nature Physics, 114, 848–854.
- Davì V, Tanimoto H, Ershov D, Haupt A, De Belly H, Le Borgne R, Couturier E, Boudaoud#A and Minc#, (2018) “Mechanosensation Dynamically Coordinates Polar Growth and Cell Wall Assembly to Promote Cell Survival” Developmental Cell, 45, 2, p170–182.
- Pierre A, Sallé J, Wühr M, Minc N., (2016) “Generic Theoretical Models to Predict Division Patterns of Cleaving Embryos.” Developmental Cell. 39(6):667-682
- Bonazzi* D, Julien* JD, Seddiki R, Romao M, Piel M, Boudaoud#A, Minc# N, (2014) “Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics.” Developmental Cell, 28 (5):534-546
- Minc#, Burgess, D. and Chang F. (2011), “Influence of cell geometry on division plane positioning” Cell., 144 (3): 414-426.
Publications
Preprints
Daria Bonazzi (2015)
Anaëlle Pierre (2017)
Valeria Davì (2018)
Héliciane Palenzuela (2020)
Jing Xie (2022)
- Guillaume Romet-Lemonne (IJM)
- Delphine Delacour (IJM)
- Aki Kimura (NIG, Japan)
- Yohanns Bellaiche (Institut Curie, Paris)
- CP Heisenberg (IST austria)
- Martin Wühr (Princeton University)
- Dan Levy (U. Wyoming).
- Arezki Boudaoud (Ecole Polytechnique)
- Etienne Couturier (MsC, Paris)
- Yolanda Sanchez (U. Cordoba)
- Miguel Peñalva (U. Madrid).
- La Ligue Contre le Cancer
- Fondation Bettencourt-Schueller
- European Research Council
- Mairie De Paris; ANR
- Fondation de la Recherche Médicale
- FungiBrain (European ITN) FP7.
20/11/2024: Impulscience Prize 2024 awarded by the Bettencourt Schueller Foundation
24/04/2024 : Mécanobiologie : la pression créatrice, contribution de l’équipe Minc à la publication d’un article dans le journal du CNRS
The Minc Lab is recruiting postdocs: