Neurobiologie développementale comparée
Nikos KONSTANTINIDES
In the lab of Comparative Developmental Neurobiology, our goal is to understand how the impressive diversity of neurons that can be found in the animal brain develops during the hours, days, or weeks of embryonic development and how it has evolved over the millions of years of evolution. We use single-cell sequencing and Drosophila powerful genetic tools to characterize complex neurodevelopmental mechanisms and understand how they have evolved.
Keywords :Neuro-evo-devo, Drosophila melanogaster, Neuronal development, Evolution of developmental mechanisms, Visual system, Single-cell sequencing
+33 (0)1 57 27 70 47 Contact @nkonst4.bsky.social www.konstantinides-lab.com
The question that drives our research
Neurons are the most diverse cell type in the animal body in terms of morphology, physiology, molecular identity, activity, function, etc. In the lab, we try to understand how this impressive neuronal diversity is being generated. We study this in two different timescales, first in the few hours, days, or months of embryonic development, and second over the millions of years of animal evolution.
In other words, we want to answer :
- How does a neuron acquire a neuronal type identity and how does it implement it?
- How do neurons that are tightly interconnected in circuits evolve to output new behaviours?
Our model
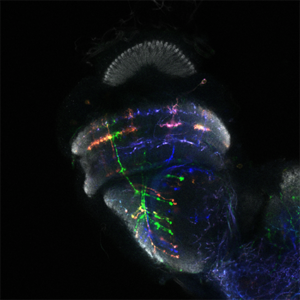
We are using the insect visual system as a model (Figure 1). Insects inhabit diverse environments with different visual landscapes and stimuli. While preserving the overall organisation of their visual system (optic lobes), they have adapted it to their environment. Thus, insect optic lobes represent an ideal system to address how neuronal systems evolve to support different behaviours. Indeed, over the last 10 years, the Drosophila optic lobe has emerged as an ideal system to address these questions for four reasons: a) It is an experimentally manageable, albeit complex structure, for which we have a very comprehensive catalogue of neuronal cell types. Meticulous work from 30 years ago had identified 100 cell types in the optic lobes based solely on morphological characters. Recent work took advantage of elaborate genetic tools, as well as single-cell mRNA sequencing, to expand the number of neuronal cell types to 169, based on both morphology and molecular identity (Figure 2). Importantly, many of the optic lobe cell types are present in multiple copies in the brain (ranging from 50 to 800), which makes clustering of single-cell sequencing (scSeq) data and cluster annotation easier. b) We know the basic principles by which most of these neuronal cell types are generated during development and how they are specified. The interplay of three mechanisms is responsible for the generation of the 169 different neuronal types: spatial patterning diversifies neural stem cells based on their spatial origin, temporal patterning allows neural stem cells of different ages to sequentially give birth to different cell types; while a Notch-driven binary cell fate decision made at the time of division of the precursor, results in the generation of two different neurons. c) A detailed connectomic map of the medulla (the largest optic ganglion), as well as a thorough enumeration of the synapses between different cell types can be used to probe circuitry. Recently, an expanded EM reconstruction of the whole brain has been made available to track neurons and circuits. d) Finally, the neuronal circuits responsible for different behaviours, such as motion detection and colour vision, walking, jumping, etc are relatively well characterized. Nonetheless, studies in one species alone cannot address evolutionary questions; the Drosophila visual system is, hence, an invaluable starting point for comparative studies across insects.
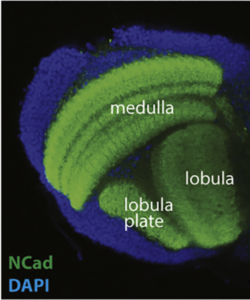
Figure 1
Our tools
We are using the Drosophila genetic arsenal (Figure 3) and sc-Seq approaches (Figure 2) as tools to answer our questions. Technological developments in scSeq approaches and in transcriptome analysis give us for the first time the opportunity to understand neuronal diversity and get access to the transcriptomes of almost all cell types in the brain. Moreover, recent research has catalogued almost all neurons in the adult brain in Drosophila melanogaster. Apart from generating atlases, significant biological insights have arisen from scSeq studies. For example, we unraveled new causal relationships between transcription factors and terminal features and found that a specific neuronal feature (e.g. neurotransmitter identity) can be the endpoint of several distinct developmental trajectories (we termed this phenotypic convergence). Similar studies in other animals have illustrated the power of scSeq to address a variety of biological questions.
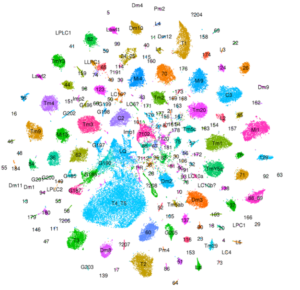
Figure 2
MORE DETAILS ABOUT THE DROSOPHILA VISUAL SYSTEM
The fly optic lobes can be divided into four neuropils: lamina, medulla, lobula and lobula plate (Figure 1). The lamina receives direct innervation from photoreceptors before relaying information to the medulla and to higher processing centers for motion detection. The lamina neurons, as well as two photoreceptors, project to the medulla. With ~40,000 cells belonging to over 100 cell types, the medulla is the most complex structure in the optic lobe. 800 ‘columns’ represent the functional units of the medulla that are processing visual information coming from one point in space in a retinotopic manner. The retinotopic organization of the medulla is somehow preserved in the lobula complex, where motion (lobula plate) and other visual features discussed in detail later (lobula), are processed.
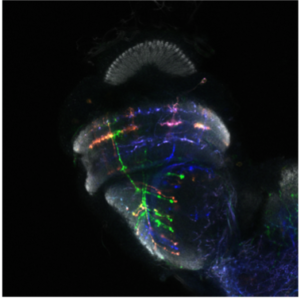 Figure 3
Figure 3
The vast majority of Drosophila optic lobe neurons are generated during the third larval stage. Two neuroepithelial structures generate neural stem cells (neuroblasts) that divide many times, self-renewing and generating an intermediate precursor, called ganglion mother cell (GMC), which divides only once to generate two neurons. Neuroblasts are capable of generating many different neuronal types using three main mechanisms. i) Temporal patterning: eleven transcription factors (TFs), Oaz, Hth, Opa, Erm, Ey, Hbn, Slp, Scro, D, BarHI, and Tll (Figure 4) are expressed in a tight temporal sequence in every neuroblast, leading to the formation of the different neuronal populations. ii) Spatial patterning: three TFs expressed in sub-regions of the neuroepithelium (Vsx, Optix, Rx) that generate the medulla modify the neuroblast identity without affecting the temporal sequence to generate locally more neuronal types. iii) Notch-driven binary cell fate decision: further neuronal diversity is achieved by Notch during the last asymmetric division of the GMC. The combination of these three mechanisms, as well as cell death, are responsible for the generation of a large proportion of the observed neuronal diversity.
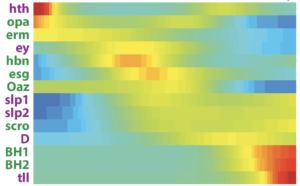
Figure 4
We are interested in understanding how these neuronal specification mechanisms have evolved and how they diversify to generate novel cell types.
Members
 Marie-Charlotte DEVAGOUROU, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Marie-Charlotte DEVAGOUROU, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Konstantina FILIPPOPOULOU, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Konstantina FILIPPOPOULOU, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Isabel Maria HOLGUERA LOPEZ, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Isabel Maria HOLGUERA LOPEZ, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Elisavet ILIOPOULOU, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Elisavet ILIOPOULOU, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Claire JULLIOT DE LA MORANDIERE, Biology engineer, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Claire JULLIOT DE LA MORANDIERE, Biology engineer, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Leo LE BOUDEC, Intern, KONSTANTINIDES LAB
Leo LE BOUDEC, Intern, KONSTANTINIDES LAB Carlos MARTIN BLANCO, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Carlos MARTIN BLANCO, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Dafni PSARAKI, Intern, KONSTANTINIDES LAB
Dafni PSARAKI, Intern, KONSTANTINIDES LAB Kyriaki Niovi RAFAILIDOU, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Kyriaki Niovi RAFAILIDOU, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Rebekah RICQUEBOURG, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Rebekah RICQUEBOURG, PhD student, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B Félix SIMON, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
Félix SIMON, Postdoctoral researcher, KONSTANTINIDES LAB+33 (0)1 57 27 70 47, room 442B
To contact a member of the team by e-mail: name.surname@ijm.fr
Marianne Malartre’s web page
Tadini, L., Younsi, L., Holguera, I., Simon, F., Courgeon, M., & Konstantinides, N. (2025). Gene expression cartography of a developing neuronal structure. bioRxiv. https://doi.org/10.1101/2025.03.30.646184
Simon, F., Holguera, I., Chen, Y.-C., Malin, J., Valentino, P., Njoo-Deplante, C., El-Danaf, R. N., Kapuralin, K., Erclik, T., Konstantinides, N., Özel, M. N., & Desplan, C. (2025). Establishment of terminal selector combinations in optic lobe neurons. bioRxiv. https://doi.org/10.1101/2024.02.05.578975
Ricquebourg, R., & Konstantinides, N. (2024). Un mécanisme temporel pour la génération de la diversité neuronale. médecine/sciences, 40(3), 251–257. https://doi.org/10.1051/medsci/2024012
Konstantinides, N., & Desplan, C. (2024). Neuronal Circuit Evolution: From Development to Structure and Adaptive Significance. Cold Spring Harbor Perspectives in Biology, a041493. https://doi.org/10.1101/cshperspect.a041493
Filippopoulou K., Konstantinides N. (2023) Evolution of patterning. The FEBS Journal 291(4):663-671. https://doi.org/10.1111/febs.16995
Konstantinides, N., Holguera, I., Rossi, A. M., Escobar, A., Dudragne, L., Chen, Y.-C., Tran, T. N., Martínez Jaimes, A. M., Özel, M. N., Simon, F., Shao, Z., Tsankova, N. M., Fullard, J. F., Walldorf, U., Roussos, P., & Desplan, C. (2022). A complete temporal transcription factor series in the fly visual system. Nature, 604(7905), 316–322. https://doi.org/10.1038/s41586-022-04564-w
Özel, M. N., Simon, F., Jafari, S., Holguera, I., Chen, Y.-C., Benhra, N., El-Danaf, R. N., Kapuralin, K., Malin, J. A., Konstantinides, N., & Desplan, C. (2021). Neuronal diversity and convergence in a visual system developmental atlas. Nature, 589(7840), 88–95. https://doi.org/10.1038/s41586-020-2879-3
Simon, F., & Konstantinides, N. (2021). Single-cell transcriptomics in the Drosophila visual system: Advances and perspectives on cell identity regulation, connectivity, and neuronal diversity evolution. Developmental Biology, 479, 107–122. https://doi.org/10.1016/j.ydbio.2021.08.001
Konstantinides, N., Kapuralin, K., Fadil, C., Barboza, L., Satija, R., & Desplan, C. (2018). Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features. Cell, 174(3), 622-635.e13. https://doi.org/10.1016/j.cell.2018.05.021
Publications
Preprint
- Toni Gabaldon (IRB, Barcelona)
- Michael Perry (University of California San Diego)

Retrouvez toutes les actualités sur : https://konstantinides-lab.com/news/
04/09/2023 : Intégration au sein du FENS-Kavli Network of Excellence


