Dynamique de la chromatine dans le développement des mammifères
MAXIM GREENBERG
DNA methylation is a stable epigenetic mark that is absolutely essential for proper mammalian development. In the Greenberg lab, the primary focus is understanding the various mechanisms and functions of DNA methylation in the context of development. We take a multidisciplinary approach, utilizing genetics, genomics, and biochemistry. We also strive to comprehend how DNA methylation interacts with other chromatin pathways to gain a holistic picture of chromatin-based gene regulation
Keywords: DNA methylation, epigenetics, chromatin, murine embryonic stem cells, development
Office: +33 (0) 1 57 27 81 20 Lab: 80 32 Contact @maxvcg.bsky.social https://www.maximgreenberglab.com/
Research aim
We strive to gain a clear understanding of the profound epigenetic consequences of DNA methylation in a window of development, which occurs in the first week of mouse embryogenesis, and the second of human, but the repercussions of which can ripple throughout life.
Background
Immediately after fertilization, mammalian genomes undergo a dramatic reshaping of the epigenome as the embryo transitions from the zygote into the pluripotent cells primed for lineage commitment. This is best exemplified by DNA methylation reprogramming, as the gametic patterns are largely erased, and the embryonic genome undergoes a wave of de novo DNA methylation. Moreover, once DNA methylation patterns are established, mechanisms faithfully maintain the mark across cell division. Thus, there is latent potential for DNA methylation deposited in the early embryo to exhibit a lifelong effect.
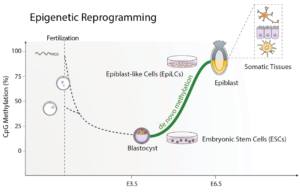
DNA methylation is a modification that is typically associated with gene repression at repetitive elements and at a minority of protein coding genes. We previously described the regulation of the Zdbf2 gene in mice, which is programmed during the de novo DNA methylation program. Challenging the paradigm, in this case DNA methylation is required for activation of a gene via antagonism of the polycomb-group of silencing proteins. If the DNA methylation fails to occur, the gene stays silent throughout life, resulting in a reduced growth phenotype.
Goals
We will try to answer a broad question: what is DNA methylation doing when it’s not at promoters? And its corollary: how do promoters stay DNA methylation free?
It has long been known that DNA methylation at CpG-dense promoters, also known as CpG island (CGI) promoters, is associated with gene silencing. However, the mammalian genome is highly methylated with the exception of CGI promoters. This means that DNA methylation is important for silencing only a small subset of protein coding genes. It is less clear what the function DNA methylation serves in intergenic regions and the bodies of transcribed genes, for example.
 One key relationship we are interested in based on our studies of Zdbf2 is the antagonism observed between DNA methylation and the polycomb silencing machinery. We are highly motivated to gain insights into the mechanistic underpinnings of DNA methylation-based gene regulation. Crucially, we want to understand the relevance of non-canonical forms of DNA methylation for development.
One key relationship we are interested in based on our studies of Zdbf2 is the antagonism observed between DNA methylation and the polycomb silencing machinery. We are highly motivated to gain insights into the mechanistic underpinnings of DNA methylation-based gene regulation. Crucially, we want to understand the relevance of non-canonical forms of DNA methylation for development.
Methods
In the Greenberg lab our playground is mouse embryonic stem cells (ESCs). ESCs offer a number of advantages for DNA methylation studies: they are resilient to mutations in DNA methyltransferase genes, they are highly amenable to CRISPR/Cas9 genome editing and genetic screens, and they can be readily harvested in high numbers for robust experiments. Importantly, we utilize a differentiation system that recapitulates the DNA methylation reprogramming that occurs in vivo. Much as we did with Zdbf2, findings we make in our cell system will be complemented in mouse models to validate the developmental importance.
We will rely on next generation sequencing (NGS) approaches to gain an in-depth view of DNA methylation regulation. This includes CUT&RUN and CUT&Tag for profiling of chromatin bound factors and targeted deep bisulfite sequencing for DNA methylation analysis. It is an exciting time in the epigenetics field with the emergence of the epigenome editing tool kit. Using CRISPR/Cas9-based targeting, we can explore with precision the function of DNA methylation at a locus-by-locus basis.

In the future, we are excited to employ genome-wide CRISPR/Cas9-based genetic screens for discovery of novel factors, as well as working with the top line proteomics platform at IJM for mass spectrometry-based approaches. We are always open to new technologies and ideas, so our methods will constantly be evolving! If you are interested in our research and want to learn more, don’t hesitate to contact us!
Members
 Elisa Carrettin, Intern, GREENBERG LAB
Elisa Carrettin, Intern, GREENBERG LAB Angélique DAVID, PhD student, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
Angélique DAVID, PhD student, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B Narjesse Hounani, Intern, GREENBERG LAB
Narjesse Hounani, Intern, GREENBERG LAB Priscillia LHOUMAUD, Researcher, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
Priscillia LHOUMAUD, Researcher, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B Marlet MORALES-FRANCO, Postdoctoral researcher, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
Marlet MORALES-FRANCO, Postdoctoral researcher, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B Julie PAULIAT, PhD student, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
Julie PAULIAT, PhD student, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B Margherita SCARPA, Biology engineer, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
Margherita SCARPA, Biology engineer, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B Teresa URLI, PhD student, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
Teresa URLI, PhD student, GREENBERG LAB+33 (0)1 57 27 81 32, room 542B
To contact a member of the team by e-mail: name.surname@ijm.fr
Greenberg M.V.C. (2021) Get Out and Stay Out: New Insights into DNA Methylation Reprogramming in Mammals. Front. Cell Dev. Biol. 8:629068
Greenberg M.V.C, Bourc’his D. (2019) The diverse roles of DNA methylation in mammalian development and disease. Nature Reviews Molecular Cell Biology 20:590-607
Greenberg M.V.C.†, Teissandier A., Walter, M., Noordermeer D., Bourc’his D†. (2019) Dynamic enhancer partitioning instructs activation of a growth-related gene during exit from naïve pluripotency. eLife 8:e44057
Greenberg M.V.C.*, Glaser J.*, Borsos M., El Marjou F., Walter M., Teissandier A., Bourc’his D. (2017) Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nature Genet. 49:110-118
F1000Prime recommendation – F1000Prime.com/726971069#eval793526035
Greenberg M.V.C., Bourc’his D. (2015) Cultural relativism: maintenance of genomic imprints in pluripotent stem cell culture systems. Curr. Op. Genet. & Dev. 31:42–49
Greenberg M.V.C.*, Deleris A.*, Hale C.J.*, Liu A., Feng S., Jacobsen S.E. (2013) Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 9(11): e1003946, 1-11
Stroud H., Greenberg M.V.C., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352-64
Ausin I.*, Greenberg M.V.C.*, Simanshu D.K.*, Hale C.J., Vashisht A.A., Simon S.A., Lee T., Feng S., Espanola S.D., Meyers B.C., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. (2012) INVOLVED IN DE NOVO 2 containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Nat. Acad. Sci. U. S. A. 109: 8374-8381
Greenberg M.V.C., Ausin I., Chan S.W.L., Cokus S.J., Cuperus J.T., Feng S., Law J.A., Chu C., Pellegrini M., Carrington J.C., Jacobsen S.E. (2011) Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics6:344-354
Deleris A.*, Greenberg M.V.C.*, Ausin I., Law R.W.Y., Moissiard G., Schubert D., Jacobsen S.E. (2010) Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA Methylation. EMBO Rep.11: 950-955
Publications
Preprint
- ANR “REMEDY”:
- Pierre-Antoine Defossez (Coordinator), Epigenetics and Cell Fate, CNRS, Université Paris Cité, Paris
- Salvatore Spicuglia (Partner), TAGC, INSERM, Aix-Marseille University, Marseille
- LabEx Who Am I? “DECODERS”:
- Benoit Palancade (Coordinator), Université Paris Cité, Institut Jacques Monod, CNRS, Paris

CNRS INSB article: Les signaux épigénétiques : comment les marques de l’ADN façonnent le plan de la vie
Le prix Georges Brahms de la Fondation CNRS attribué à Priscillia Lhoumaud


