Division cellulaire et reproduction
Julien DUMONT
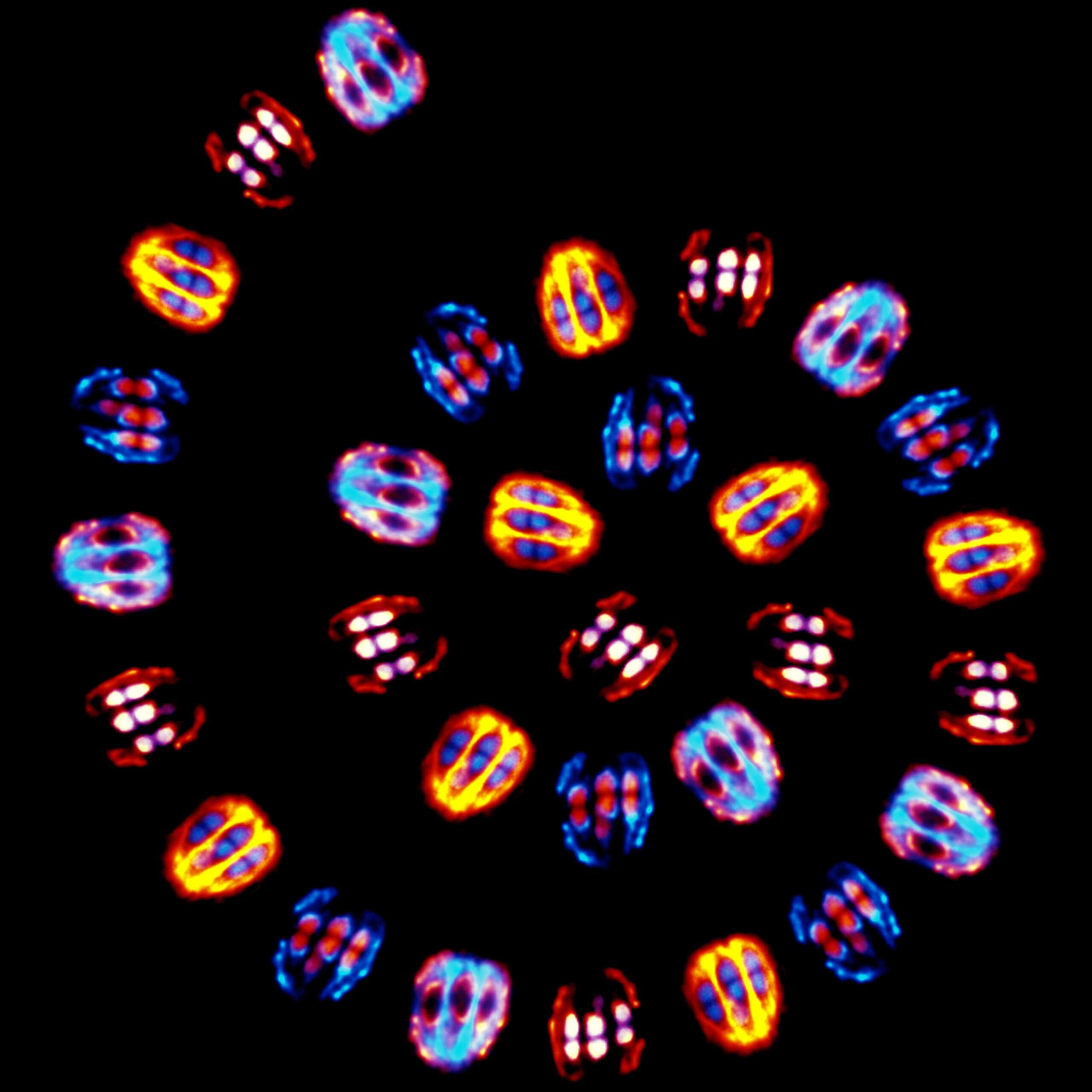 During cell division, the chromosomes, which carry the genetic information, are distributed into two equal complements between the two daughter cells. During development, aneuploidy, which corresponds to the presence of an incorrect number of chromosomes, leads to the formation of embryos that are generally non-viable or with severe developmental defects. Our project aims to study the mechanisms that ensure the formation of oocytes and embryos containing the correct number of chromosomes. For this, we study the assembly mechanisms and the function of the cellular machinery allowing the correct distribution of chromosomes in meiosis and mitosis. Our approach is multidisciplinary and relies on state-of-the-art genetic, biochemical, cellular and microscopic approaches.
During cell division, the chromosomes, which carry the genetic information, are distributed into two equal complements between the two daughter cells. During development, aneuploidy, which corresponds to the presence of an incorrect number of chromosomes, leads to the formation of embryos that are generally non-viable or with severe developmental defects. Our project aims to study the mechanisms that ensure the formation of oocytes and embryos containing the correct number of chromosomes. For this, we study the assembly mechanisms and the function of the cellular machinery allowing the correct distribution of chromosomes in meiosis and mitosis. Our approach is multidisciplinary and relies on state-of-the-art genetic, biochemical, cellular and microscopic approaches.
Keywords: Cell division, meiosis/mitosis, cytoskeleton, chromosome segregation, embryonic development.
+33 (0)1 57 27 80 49 Contact
Approximately 15% of couples are confronted with an infertility problem. The potential causes are the poor quality of the germ cells or gametes (sperm and oocyte), or an impaired embryo development. A major challenge in reproductive biology is to understand the mechanisms of these defects.
During cell division, the chromosomes, carriers of genetic information, are divided into two equal sets between the daughter cells. The faithful distribution of chromosomes is a fundamental element of the genetic stability of cells and organisms. Aneuploidy, which corresponds to the presence of an incorrect number of chromosomes, leads to the formation of embryos that are generally non-viable or show severe developmental defects. Our project aims to study the mechanisms that ensure the formation of oocytes and embryos with the correct number of chromosomes, which is essential for reproduction.
We study the mechanisms of assembly and the functioning of the cellular machinery allowing the correct distribution of chromosomes in meiosis and mitosis. This machinery is composed of a spindle formed by microtubules (green in figure 1), some of which contact chromosomes (magenta). Interactions between spindle microtubules and chromosomes are central to the process of chromosome segregation and are regulated differently in meiosis and in mitosis.
Our approach is multidisciplinary and relies on state-of-the-art genetic, biochemical, cellular and microscopic tools. Our favorite model system is the nematode worm Caenorhabditis elegans, which shares the vast majority of its genes and key mechanisms of reproduction and development with mammals. We also study “non-model” nematodes to develop an evolutionary understanding of the mechanisms of cell division in the context of reproduction.
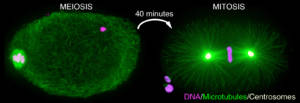
Figure 1: Immunofluorescent labeling of an oocyte meiotic spindle (left) and a zygote mitotic spindle (right) in C. elegans. Microtubules are in green, chromosomes in magenta and the two centrosomes that form the poles of the spindle in mitosis are in yellow. Note the absence of centrosomes at the poles of the meiotic spindle and its characteristic “barrel” shape.
Axis 1/ Meiotic divisions of oocyte:
In mitosis, spindle microtubules are assembled from centrosomes (Figure 1, yellow). In oocytes, centrosomes are eliminated early on during meiosis. Thus, spindle assembly in oocytes involves atypical mechanisms that are not well understood. We have developed an in utero microscopy approach on live nematodes that allowed us to describe the key steps of acentrosomal spindle formation in the C. elegans oocyte (Video 1).
In order to determine the actors involved, we combine this confocal imaging method with a systematic loss-of-function approach of microtubule-associated proteins and microtubule motors. We also use an in vitro Total Internal Reflection Microscopy (TIRF) approach on recombinant proteins and individual microtubules (Figure 2). Our goal is to determine the network of actors required for spindle formation in the absence of centrosome, as well as to highlight the functional interactions between these actors.
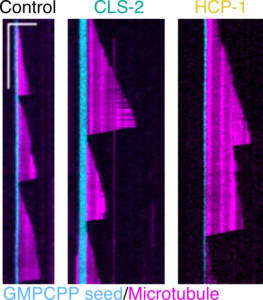
Figure 2
Axis 2/ Chromosome segregation in the oocyte:
Acentrosomal spindles have a characteristic “barrel” shape (Figure 1), with rounded poles, due to the absence of astral microtubules (microtubules that normally connect the spindle poles to the cell cortex during mitosis). This atypical shape induces specific mechanisms of meiotic chromosome segregation. Using a multimodal microscopy approach, combining live optical microscopy of C. elegans oocytes, laser ablation on live oocytes and tomographic electron microscopy, we have demonstrated an original mechanism of chromosome segregation relying on “pushing” forces exerted by microtubules (Video 2 and Figure 3). We combine the different microscopy approaches with targeted loss-of-function of key players to determine the molecular mechanisms.
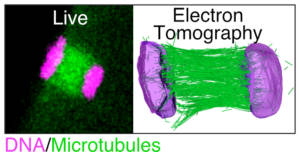
Figure 3
We have also shown that the “pushing” segregation is not strictly dependent on kinetochores, multiprotein complexes that assemble on chromosomes to allow microtubule binding. Nevertheless, these are important to ensure the fidelity of the segregation mechanism and to avoid oocyte aneuploidy. We have recently developed a method of live microscopy of two fluorescence channels simultaneously with fast Z-scan (Figure 4).
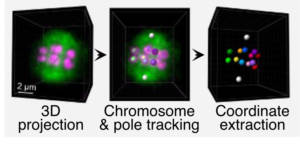
Figure 4
This method allows us to acquire the full volume of the meiotic spindle and segregating chromosomes during the entire first meiotic division. This allows us to follow the position of each chromosome individually during the orientation, alignment and segregation steps on the meiotic spindle. This approach allows us to perform a quantitative analysis of the function of candidate proteins and to determine their importance for chromosome segregation in the C. elegans oocyte.
Axis 3/ Evolution of the mechanisms of meiotic division in nematodes:
Defects in chromosome segregation during meiosis can lead to aneuploidy of gametes and embryos, which is a major cause of birth defects and spontaneous abortions in humans. However, although essential for the reproduction of multicellular organisms and thus for the continuity of species, the key principles governing meiosis, and in particular meiotic chromosome segregation in oocytes, are still poorly understood. One of the main reasons for this lack of knowledge is the diversity of molecular scenarios that have been adopted during evolution to regulate chromosome segregation, particularly in oocytes. Indeed, although meiosis is highly conserved in eukaryotes, deviations from the “norm” are ubiquitous and may provide important information about the evolutionary significance of meiotic mechanisms. It is therefore of utmost importance to appreciate the true diversity of meiotic features in nature, considering both well-characterized model organisms (such as C. elegans) and non-model organisms. Nematodes represent an ideal situation in this respect: their remarkable adaptability to various ecological niches has been accompanied by extreme plasticity in genome organization but also in reproductive and meiotic strategies. We have recently started the analysis of oocyte meiosis in non-model nematode species with specific and atypical characteristics that may have an impact on meiotic chromosome segregation (Video 3).
Axis 4/ The first mitotic division of the c.elegans zygote:
In the C. elegans zygote, during mitosis, microtubules emanating from the spindle poles contact either the embryonic cortex (astral microtubules) or the chromosomes via the kinetochores (kinetochore microtubules). Pulling forces exerted by astral microtubules are transmitted to chromosomes by kinetochore microtubules allowing the segregation of sister chromatids in anaphase (Video 4).
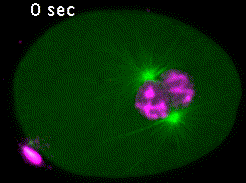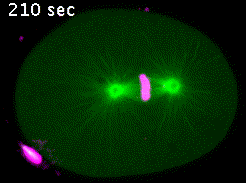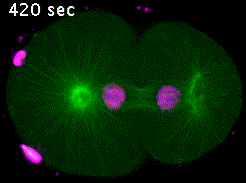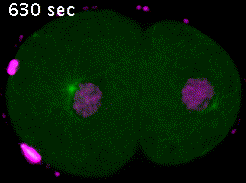
Kinetochores are multiprotein complexes composed of several dozen proteins. Among these, the kinase BUB-1, the HCP-½ proteins, and the microtubule-associated protein CLS-2, form a module involved in the control of kinetochore microtubule dynamics. We have highlighted the roles of this module in the correct alignment and attachment of chromosomes in metaphase, and in the formation of the anaphase central spindle. We continue to analyze the different functions of the BUB-1/HCP-½/CLS-2 module during the first mitotic division of the C. elegans zygote. For this purpose, we are developing a targeted mutagenesis approach coupled to confocal microscopy on living embryos. This approach allows us to perform a quantitative analysis of the role of the different actors through the various stages of mitosis (Figure 5).
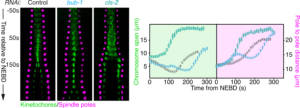
Figure 5
Members
 Noemie FHIMA, Biology engineer, DUMONT LAB+33 (0)1 57 27 80 49, room 402B
Noemie FHIMA, Biology engineer, DUMONT LAB+33 (0)1 57 27 80 49, room 402B Laura GUEDON, Biology engineer, DUMONT LAB
Laura GUEDON, Biology engineer, DUMONT LAB Leo LENHARDT, PhD student, DUMONT LAB
Leo LENHARDT, PhD student, DUMONT LAB Moc Linh Huong LUU, Intern, DUMONT LAB
Moc Linh Huong LUU, Intern, DUMONT LAB Gilliane MATON, Assistant Professor, DUMONT LAB+33 (0)1 57 27 80 47, room 402B
Gilliane MATON, Assistant Professor, DUMONT LAB+33 (0)1 57 27 80 47, room 402B
To contact a member of the team by e-mail: name.surname@ijm.fr

Dias Maia Henriques AM, Davies T, Dmitrieff S, Minc N, Canman JC, Dumont J# & Maton G#. Mechanical coordination between anaphase A and B drives asymmetric chromosome segregation. J Cell Biol doi: 10.1083/jcb.202505038 (2026)
El Mossadeq L, Bellutti L, Le Borgne R, Canman JC, Pintard L, Verbavatz J-M, Askjaer P & Dumont J. An interkinetic envelope surrounds chromosomes between meiosis I and II in C. elegans oocytes. J Cell Biol doi: 10.1083/jcb.202403125 (2025)
Belluti L, Macaisne N, El Mossadeq L, Ganeswaran T, Canman JC & Dumont J. Regulation of outer kinetochore assembly during meiosis I and II by CENP-A and KNL-2/M18BP1 in C. elegans oocytes. Current Biology S0960-9822(24)01215-6 doi: 10.1016/j.cub.2024.09.004 (2024)
Perrier A, Guiglielmoni N, Naquin D, Gorrichon K, Thermes C, Lameiras S, Dammermann A, Schiffer PH, Brunstein M, Canman JC & Dumont J. Maternal inheritance of functional centrioles in two parthenogenic nematodes. Nat Comm 15(1):6042. doi: 10.1038/s41467-024-50427-5 (2024)
Gareil N*, Gervais A*, Macaisne N, Chevreux G, Canman JC, Andreani J & Dumont J. An unconventional TOG domain is required for CLASP localization. Current Biology, 33(16):3522-3528.e7 doi: 10.1016/j.cub.2023.07.009 (2023)
Pitayu-Nugroho L*, Aubry M*, Laband K, Geoffroy H, Ganeswaran T, Primadhanty A, Canmam JC & Dumont J. Kinetochore component function in C. elegans oocytes revealed by 4D tracking of holocentric chromosomes. Nat Comm, 14, 4032. doi: 10.1038/s41467-023-39702-z (2023)
Macaisne N*, Bellutti L*, Laband K*, Edwards F*, Pitayu-Nugroho L, Gervais A, Ganeswaran T, Geoffroy H, Maton G, Canman JC, Lacroix B & Dumont J. Synergistic stabilization of microtubules by BUB-1, HCP-1 and CLS-2 controls microtubule pausing and meiotic spindle assembly. eLife, 12:e82579. doi: 10.7554/eLife.82579 (2023)
Edwards F, Maton G, Gareil N, Canman JC & Dumont J. BUB-1 promotes amphitelic chromosome biorientation via multiple activities at the kinetochore. eLife 2018;7:e40690 doi: 10.7554/eLife.40690 (2018)
Lacroix B, Letort G, Pitayu L, Sallé J, Stefanutti M, Maton G, Ladouceur AM, Canman JC, Maddox PS, Maddox AS, Minc N, Nédélec F & Dumont J. Microtubule dynamics scale with cell size to set spindle length and assembly timing. Dev. Cell, 45, (4):496-511 doi: 10.1016/j.devcel.2018.04.022 (2018) Highlighted in Dev Cell 45:421-423, (2018) F1000: https://f1000-com.insb.bib.cnrs.fr/prime/733268901#eval793546852
Laband K, Le Borgne R, Edwards F, Stefanutti M, Canman JC, Verbavatz J-M & Dumont J. Chromosome segregation occurs by microtubule pushing in oocytes. Nat Commun, 14;8(1):1499 doi: 10.1038/s41467-017-01539-8 (2017)
Gigant E*, Stefanutti M*, Laband K, Gluszek-Kustusz A, Edwards F, Maton G, Lacroix B, Canman JC, Welburn J & Dumont J. Inhibition of ectopic microtubule assembly by the kinesin-13 KLP-7MCAK prevents chromosome segregation and cytokinesis defects in oocytes. Development, 144(9):1674-1686, doi: 10.1242/dev.147504 (2017)
Maton G*, Edwards F*, Lacroix B, Stefanutti M, Laband K, Lieury T, Kim T, Espeut J, Canman JC, & Dumont J. Kinetochore components are required for central spindle assembly. Nat Cell Biol, 17(5):697-705 doi: 10.1038/ncb3150 (2015) Dispatched in Current Biology 25, R554–R557, (2015)
Dumont J, Oegema K & Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol,12(9):894-901 doi: 10.1038/ncb2093 (2010)
Publications
2026
Dias Maia Henriques AM, Davies T, Dmitrieff S, Minc N, Canman JC, Dumont J# & Maton G#. Mechanical coordination between anaphase A and B drives asymmetric chromosome segregation. J Cell Biol doi: 10.1083/jcb.202505038 (2026)
2025
El Mossadeq L, Bellutti L, Le Borgne R, Canman JC, Pintard L, Verbavatz J-M, Askjaer P & Dumont J. An interkinetic envelope surrounds chromosomes between meiosis I and II in C. elegans oocytes. J Cell Biol doi: 10.1083/jcb.202403125 (2025)
Xie J, Najafi J, Nommick A, Lederer L, Salle J, Dmitrieff S, Lacroix B, Dumont J & Minc N. Cell shape modulates mitotic spindle positioning forces via intracellular hydrodynamics. Current Biology doi: 10.1016/j.cub.2024.11.055 (2025)
2024
El Mossadeq L, Bellutti L, Le Borgne R, Canman JC, Pintard L, Verbavatz J-M, Askjaer P & Dumont J. An interkinetic envelope surrounds chromosomes between meiosis I and II in C. elegans oocytes. J Cell Biol (accepted, 2024)
Xie J, Najafi J, Nommick A, Lederer L, Salle J, Dmitrieff S, Lacroix B, Dumont J. & Minc N. Cell shape modulates mitotic spindle positioning forces via intracellular hydrodynamics. Current Biology (accepted, 2024)
Belluti L, Macaisne N, El Mossadeq L, Ganeswaran T, Canman JC & Dumont J. Regulation of outer kinetochore assembly during meiosis I and II by CENP-A and KNL-2/M18BP1 in C. elegans oocytes. Current Biology S0960-9822(24)01215-6 doi: 10.1016/j.cub.2024.09.004 (2024)
Perrier A, Guiglielmoni N, Naquin D, Gorrichon K, Thermes C, Lameiras S, Dammermann A, Schiffer PH, Brunstein M, Canman JC & Dumont J. Maternal inheritance of functional centrioles in two parthenogenic nematodes. Nat Comm 15(1):6042. doi: 10.1038/s41467-024-50427-5 (2024)
Connors CQ, Martin SL, Dumont J, Shirasu-Hiza M, Canman JC. Cell type-specific regulation by different cytokinetic pathways in the early embryo. MicroPubl Biol. Oct 22;2024:10.17912/micropub.biology.001316. doi:10.17912/micropub.biology.001316 (2024)
Connors CQ, Mauro MS, Tristian Wiles J, Countryman AD, Martin SL, Lacroix B, Shirasu-Hiza M, Dumont J, Kasza KE, Davies TR & Canman JC. Germ fate determinants protect germ precursor cell division by restricting septin and anillin levels at the division plane. MBoC 35(7), doi: 10.1101/2023.11.17.566773 (2024)
2023
Gareil N*, Gervais A*, Macaisne N, Chevreux G, Canman JC, Andreani J & Dumont J. An unconventional TOG domain is required for CLASP localization. Current Biology, 33(16):3522-3528.e7 doi: 10.1016/j.cub.2023.07.009 (2023)
Pitayu-Nugroho L*, Aubry M*, Laband K, Geoffroy H, Ganeswaran T, Primadhanty A, Canmam JC & Dumont J. Kinetochore component function in C. elegans oocytes revealed by 4D tracking of holocentric chromosomes. Nat Comm, 14, 4032. doi: 10.1038/s41467-023-39702-z (2023)
Macaisne N*, Bellutti L*, Laband K*, Edwards F*, Pitayu-Nugroho L, Gervais A, Ganeswaran T, Geoffroy H, Maton G, Canman JC, Lacroix B & Dumont J. Synergistic stabilization of microtubules by BUB-1, HCP-1 and CLS-2 controls microtubule pausing and meiotic spindle assembly. eLife, 12:e82579. doi: 10.7554/eLife.82579 (2023)
Lignieres L, Senecaut N, Dang T, Bellutti L, Hamon M, Terrier S, Legros V, Chevreux G, Lelandais G, Mege RM, Dumont J & Camadro JM. Extending the range of SLIM-labeling applications: From human cell lines in culture to Caenorhabditis elegans whole-organism labeling. J Proteome Research, 22(3):996-1002. doi: 10.1021/acs.jproteome.2c00699 (2023)
Rocha H, Simões PA, Budrewicz J, Lara-Gonzalez P, Carvalho AX, Dumont J, Desai A, Gassmann R. Nuclear-enriched protein phosphatase 4 ensures outer kinetochore assembly prior to nuclear dissolution. J Cell Biol, 222(3):e202208154 doi: 10.1083/jcb.202208154 (2023)
2022
Hirsch S, Edwards F, Shirasu-Hiza M, Dumont J & Canman JC. Functional midbody assembly in the absence of a central spindle. J Cell Biol, 221(3):e202011085 doi: 10.1083/jcb.202011085 (2022)
2019
Cabral G, Laos T, Dumont J, & Dammermann A. Differential requirements for centrioles in mitotic centrosome growth and maintenance. Dev. Cell, 50, (3):355-366 doi: 10.1016/j.devcel.2019.06.004 (2019)
2018
Edwards F, Maton G, Gareil N, Canman JC & Dumont J. BUB-1 promotes amphitelic chromosome biorientation via multiple activities at the kinetochore. eLife 2018;7:e40690 doi: 10.7554/eLife.40690 (2018)
Hirsch SM, Sundaramoorthy S, Davies T, Zhuravlev Y, Waters JC, Shirasu-Hiza M, Dumont J & Canman JC. FLIRT: Fast Local InfraRed Thermogenetics for subcellular control of protein function. Nature Methods 2018 Nov;15(11):921-923 doi: 10.1038/s41592-018-0168-y (2018)
Davies T, Kim HX, Romano Spica N, Lesea-Pringle BJ, Dumont J, Shirasu-Hiza M & Canman JC. Cell-intrinsic and extrinsic mechanisms promote cell-type specific cytokinetic diversity. eLife 2018;7:e36204 doi: 10.7554/eLife.36204 (2018)
Lacroix B, Letort G, Pitayu L, Sallé J, Stefanutti M, Maton G, Ladouceur AM, Canman JC, Maddox PS, Maddox AS, Minc N, Nédélec F & Dumont J. Microtubule dynamics scale with cell size to set spindle length and assembly timing. Dev. Cell, 45, (4):496-511 doi: 10.1016/j.devcel.2018.04.022 (2018) Highlighted in Dev Cell 45:421-423, (2018) F1000: https://f1000-com.insb.bib.cnrs.fr/prime/733268901#eval793546852
Berto A, Yu J, Morchoisne-Bolhy S, Bertipaglia C, Vallee R, Dumont J, Ochsenbein F, Guerois R & Doye V. Disentangling the molecular determinants for Cenp-F localization to nuclear pores and kinetochores. EMBO Report 19(5). doi: 10.15252/embr.201744742 (2018)
2017
Laband K, Le Borgne R, Edwards F, Stefanutti M, Canman JC, Verbavatz J-M & Dumont J. Chromosome segregation occurs by microtubule pushing in oocytes. Nat Commun, 14;8(1):1499 doi: 10.1038/s41467-017-01539-8 (2017)
Luscan R, Mechaussier S, Paul A, Tian G, Gérard X, Loundon N, Defoort-Delhemmes S, Audo I, Dumont J, Goudin N, Garfa-Traoré M, Bras M, Pouliet A, Attié-Bittach T, Boddaert N, Lyonnet S, Kaplan J, Cowan NJ, Rozet J-M, Marlin S & Perrault I. Mutations in TUBB4B cause a distinctive sensorineural disease. Am J Hum Genet, 101(6):1006-1012 doi: 10.1016/j.ajhg.2017.10.010 (2017)
Martino L, Morchoisne-Bolhy S, Cheerambathur D, Van Hove L, Dumont J, Joly N, Desai A, Doye V & Pintard L. Channel nucleoporins recruit PLK-1 to nuclear pore complexes to direct nuclear envelope breakdown in C. elegans. Dev. Cell, 43, (2):157-171 doi: 10.1016/j.devcel.2017.09.019 (2017)
Gigant E*, Stefanutti M*, Laband K, Gluszek-Kustusz A, Edwards F, Maton G, Lacroix B, Canman JC, Welburn J & Dumont J. Inhibition of ectopic microtubule assembly by the kinesin-13 KLP-7MCAK prevents chromosome segregation and cytokinesis defects in oocytes. Development, 144(9):1674-1686, doi: 10.1242/dev.147504 (2017)
Zhuravlev Y, Hirsch S, Jordan S, Dumont J, Shirasu-Hiza M & Canman JC. CYK-4 regulates Rac, but not Rho, during cytokinesis. MBoC, 28(9):1258-1270 doi: 10.1091/mbc.E17-01-0020 (2017)
Sundaramoorthy S, Garcia BA, Hirsch S, Park JH, Davies T, Dumont J, Shirasu-Hiza M, Kummel A & Canman JC. Low efficiency upconversion nanoparticles for high-resolution coalignment of near-infrared and visible light paths on a light microscope. ACS Appl. Mater. Interfaces, 9 (9), pp 7929-7940 doi: 10.1021/acsami.6b15322 (2017)
Review
2024
Lacroix B & Dumont J. Spatial and Temporal Scaling of Microtubules and Mitotic Spindles. Cells, 11(2):248 doi: 10.3390/cells11020248 (2022)
Book chapter
2024
Dumont J & Maton G. 4D microscopy and tracking of chromosomes and the spindle in C. elegans early embryos. Methods Mol Biol (2024)
Chenevert J, Robert MLV, Sallé J, Cacchia S, Lorca T, Castro A, McDougall A, Minc N, Castagnetti S, Dumont J & Lacroix B. Measuring Mitotic Spindle and Microtubule Dynamics in Marine Embryos and Non-model Organisms. Methods Mol Biol, 2024:2740:187-210 doi: 10.1007/978-1-0716-3557-5_12 (2024)
Laband K, Lacroix B, Edwards F, Canman JC & Dumont J. Live imaging of C. elegans oocytes and early embryos. Methods Cell Biol, 145:217-236 doi: 10.1016/bs.mcb.2018.03.025 (2018)
Davies T, Sundaramoorthy S, Jordan SN, Shirasu-Hiza M, Dumont J & Canman JC. Using fast-acting temperature sensitive mutants to study cell division in C. elegans. Methods Cell Biol, 137:283-306 doi: 10.1016/bs.mcb.2016.05.004 (2017)
- Kimberley Laband : 2017
- Frances Edwards : 2018
- Layla El Mossadeq : 2022
- Mélanie Aubry : 2023

- Aurélien Perrier : 2023

- Jean-Marc Verbavatz (IJM)
- Lionel Pintard (IJM)
- Julie C. Canman (Columbia University, NYC, USA)
- Marie Delattre, (ENS, France) ; Peter Askjaer, (CABD, Spain)
- Peter Meister, (University of Bern, Switzerland)
- European Research Council
- Mairie De Paris
- Fondation pour la Recherche Médicale
- Agence Nationale de la Recherche
12/08/2025: The scientific article “An interkinetic envelope is assembled during oocyte meiosis” published by Layla El Mossadeq et al. (Dumont Lab) is one of the 13 most influential articles of the year that have attracted exceptional interest from readers of The Journal of Cell Biology.


