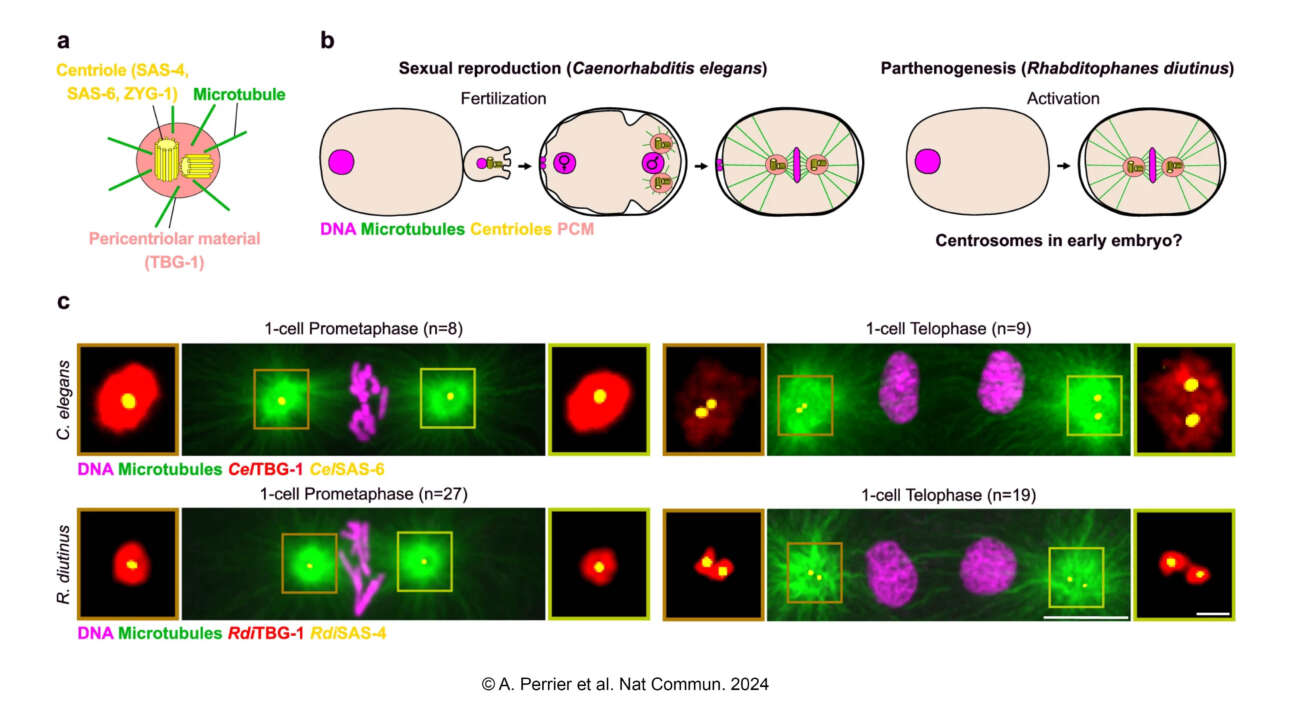
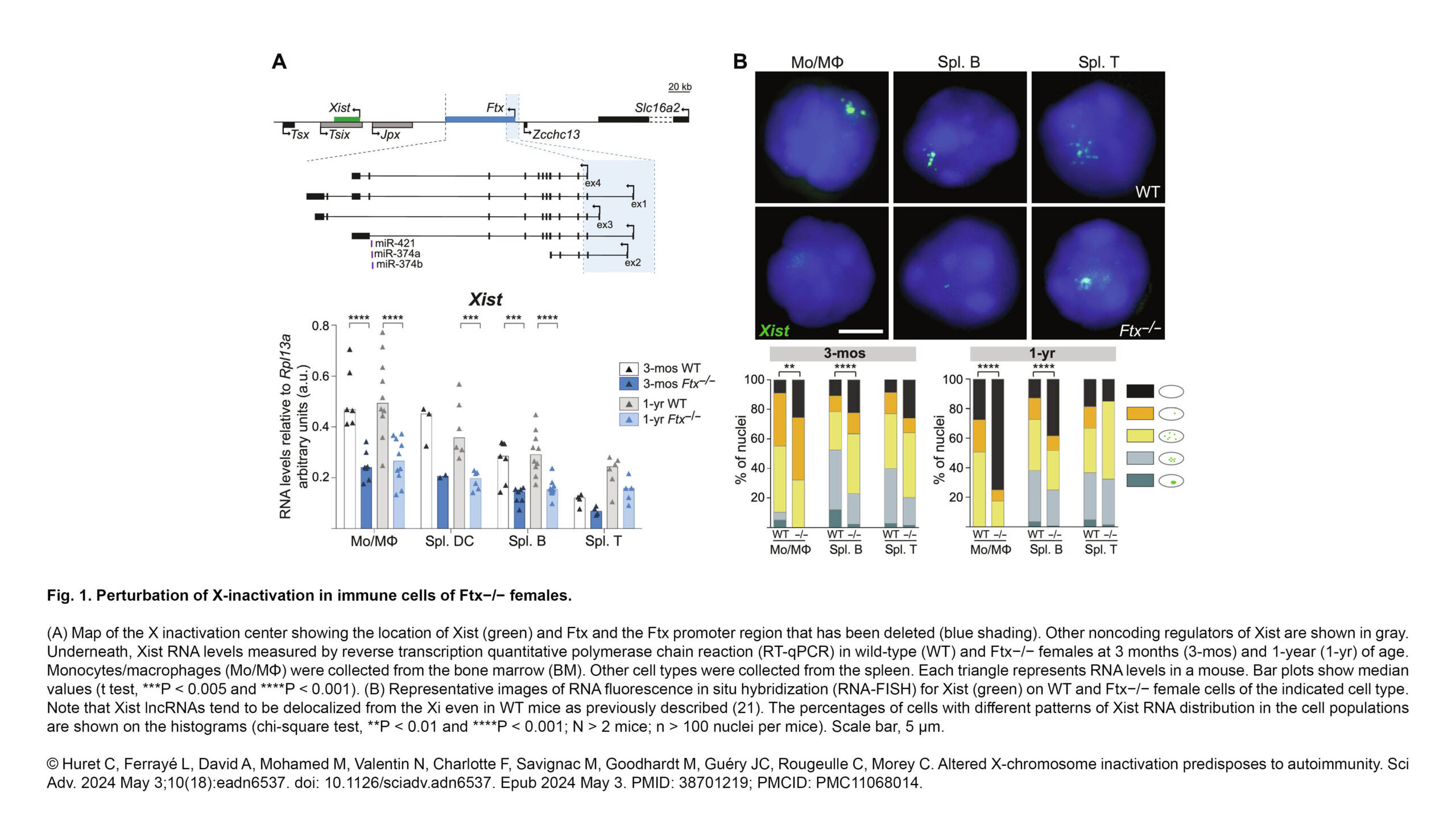
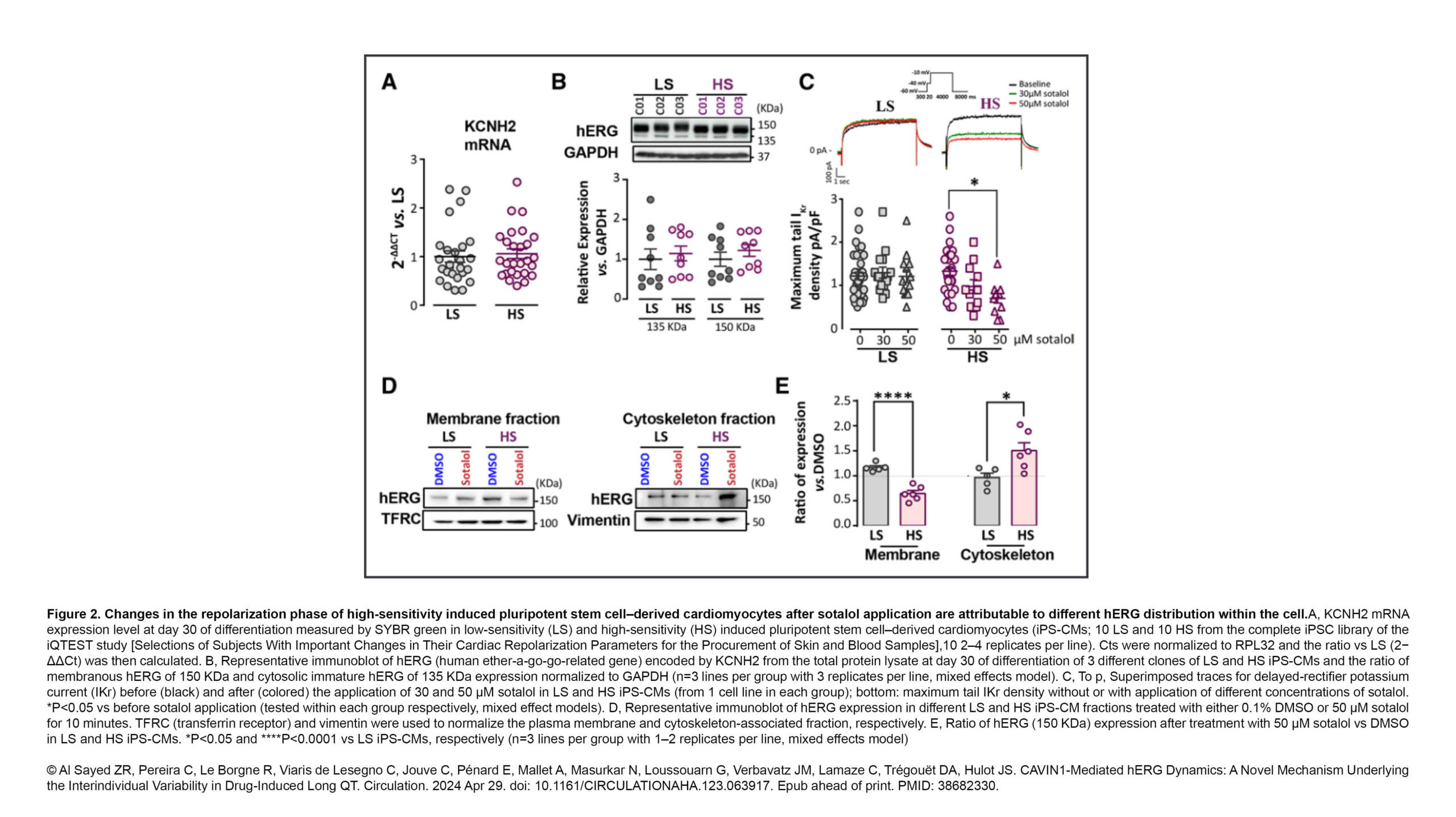
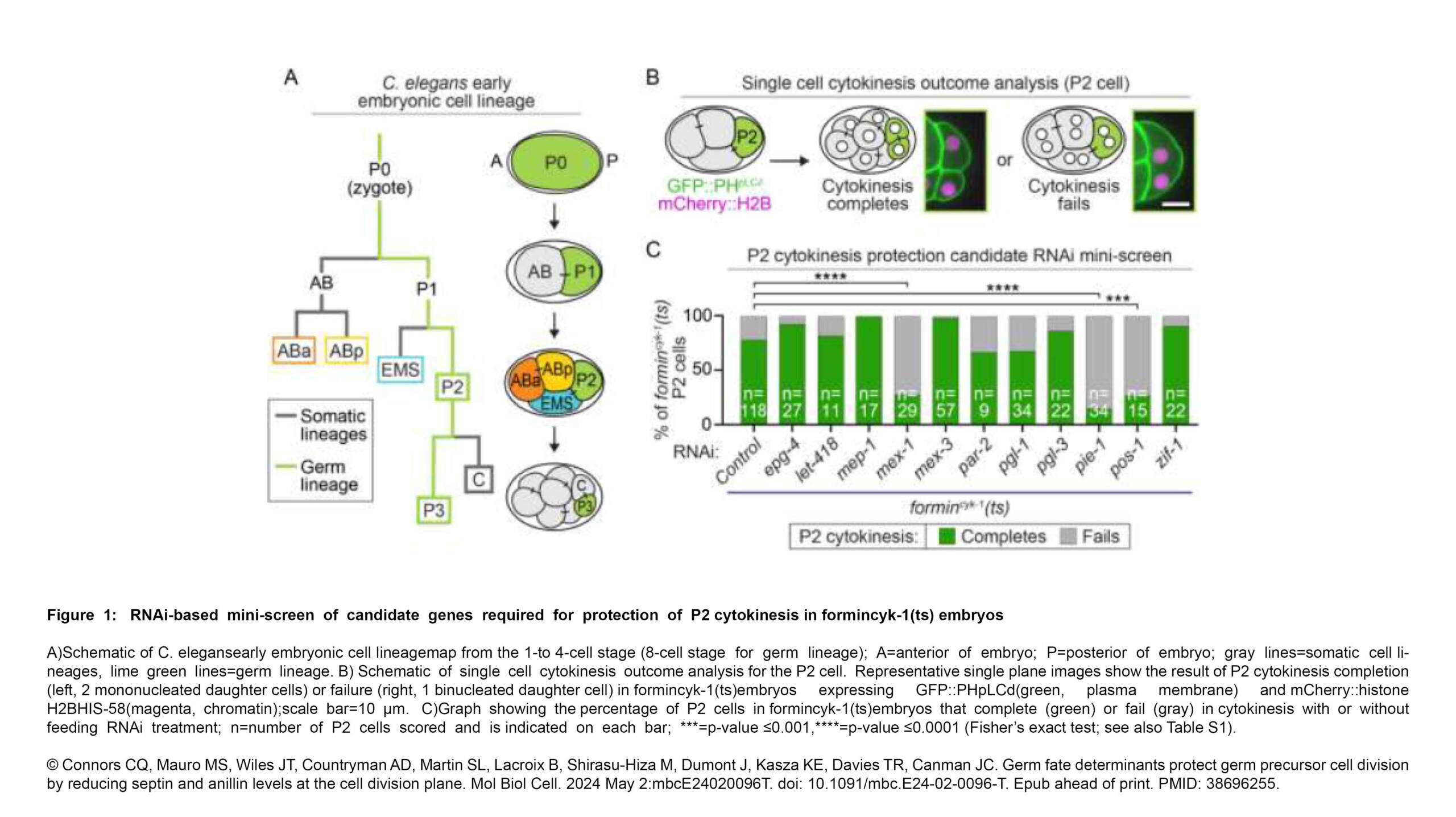
L'équipe Dumont a publié un nouvel article dans Nature Communication :
Maternal inheritance of functional centrioles in two parthenogenetic nematodes
Résumé :
Centrioles are the core constituent of centrosomes, microtubule-organizing centers involved in directing mitotic spindle assembly and chromosome segregation in animal cells. In sexually reproducing species, centrioles degenerate during oogenesis and female meiosis is usually acentrosomal.…