Neurobiologie développementale comparée
Nikos KONSTANTINIDES
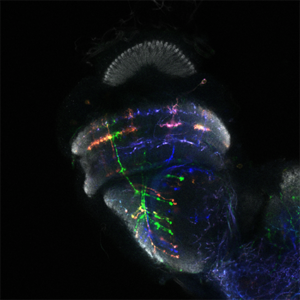
Dans le laboratoire de neurobiologie développementale comparée, notre objectif est de comprendre comment l’impressionnante diversité neuronale que l’on trouve dans le cerveau animal se développe pendant les heures, les jours ou les semaines du développement embryonnaire et comment elle a évolué au cours des millions d’années d’évolution. Nous utilisons le séquençage unicellulaire et les puissants outils génétiques de la drosophile pour caractériser les mécanismes complexes du neurodéveloppement et comprendre comment ils ont évolué.
Mots-clés : Neuro-evo-devo, Drosophila melanogaster, Neuronal development, Evolution of developmental mechanisms, Visual system, Single-cell sequencing
+33 (0)157278145 nikos.konstantinides(at)ijm.fr @nkonst4 www.konstantinides-lab.com
La question qui motive notre recherche
Les neurones constituent le type cellulaire le plus diversifié du corps animal en termes de morphologie, de physiologie, d’identité moléculaire, d’activité, de fonction, etc. Au laboratoire, nous essayons de comprendre comment cette impressionnante diversité neuronale est générée. Nous l’étudions à deux échelles de temps différentes, d’abord pendant les quelques heures, jours ou mois du développement embryonnaire, puis au cours des millions d’années de l’évolution animale.
En d’autres termes, nous voulons répondre aux questions suivantes :
- Comment un neurone acquiert-il une identité de type neuronal et comment la met-il en œuvre ?
- Comment les neurones, qui sont étroitement interconnectés dans des circuits, évoluent-ils pour produire de nouveaux comportements ?
Notre modèle
Nous utilisons le système visuel des insectes comme modèle (Figure 1). Ces derniers vivent dans des environnements variés avec des paysages visuels et des stimuli différents. Tout en conservant l’organisation générale de leur système visuel (lobes optiques), ils l’ont adapté à leur environnement. Ainsi, les lobes optiques des insectes représentent un système idéal pour étudier comment les systèmes neuronaux évoluent pour soutenir différents comportements. En effet, au cours des dix dernières années, le lobe optique de la drosophile est apparu comme un système idéal pour aborder ces questions, et ce, pour quatre raisons : a) Il s’agit d’une structure complexe, mais accessible expérimentalement, pour laquelle nous disposons d’un catalogue très complet de types de cellules neuronales. Des travaux méticuleux, réalisés il y a 30 ans, avaient permis d’identifier 100 types de cellules dans les lobes optiques sur la seule base de caractères morphologiques. Des travaux récents ont tiré parti d’outils génétiques élaborés ainsi que du séquençage de l’ARNm en cellules uniques pour étendre ce nombre de types de cellules neuronales à 169, s’appuyant sur leur morphologie et identité moléculaire (figure 2). Il est important de noter que de nombreux types cellulaires du lobe optique sont présents en plusieurs exemplaires dans le cerveau (de 50 à 800), ce qui facilite le regroupement des données de séquençage de cellules individuelles (scSeq) et l’annotation des groupes. b) Nous connaissons les principes de base selon lesquels la plupart de ces types de cellules neuronales sont générés au cours du développement et comment ils sont spécifiés. L’interaction de trois mécanismes est responsable de la génération des 169 différents types de neurones : la structuration spatiale diversifie les cellules souches neurales en fonction de leur origine spatiale, la structuration temporelle permet aux cellules souches neurales d’âges différents de donner successivement naissance à différents types de cellules, tandis qu’une décision binaire sur le destin cellulaire, pilotée par Notch et prise au moment de la division du précurseur, entraîne la génération de deux neurones différents. c) Une carte connectomique détaillée de la medulla (le plus grand ganglion optique), ainsi qu’une énumération complète des synapses entre les différents types cellulaires peuvent être utilisées pour sonder les circuits. Récemment, une reconstruction étendue par microscopie électronique du cerveau entier a été mise à disposition pour suivre les neurones et les circuits. d) Enfin, les circuits neuronaux responsables de différentes fonctions, telles que la détection de mouvement et la vision des couleurs, la marche, le saut, etc. sont relativement bien caractérisés. Néanmoins, les études menées sur une seule espèce ne permettent pas de répondre aux questions relatives à l’évolution ; le système visuel de la drosophile est donc un point de départ inestimable pour des études comparatives entre insectes.
Figure 1
Nos outils
Nous utilisons l’arsenal génétique de la drosophile (figure 3) et les approches sc-Seq (figure 2) comme outils pour répondre à nos questions. Les développements technologiques dans les approches sc-Seq et dans l’analyse du transcriptome nous donnent pour la première fois l’opportunité de comprendre la diversité neuronale et d’avoir accès aux transcriptomes de presque tous les types cellulaires du cerveau. De plus, des recherches récentes ont permis de cataloguer presque tous les neurones du cerveau adulte de Drosophila melanogaster. Outre la production d’atlas, les études de scSeq ont permis d’obtenir des informations biologiques importantes. Par exemple, nous avons découvert de nouvelles relations causales entre les facteurs de transcription et les caractéristiques terminales, et avons constaté qu’une caractéristique neuronale spécifique (par exemple, l’identité du neurotransmetteur) peut être le point final de plusieurs trajectoires distinctes de développement (nous avons appelé cela la convergence phénotypique). Des études similaires menées chez d’autres animaux ont illustré la capacité du scSeq à répondre à une variété de questions biologiques.
Figure 2
Plus de détails sur le système visuel de la drosophile
Les lobes optiques des mouches peuvent être divisés en quatre neuropiles : lamina, medulla, lobula et lobula plate (Figure 2). La lamina reçoit une innervation directe des photorécepteurs avant de relayer l’information à la médulla et aux centres de traitement supérieurs pour la détection des mouvements. Les neurones de la lamina, ainsi que deux photorécepteurs, projettent vers la medulla. Avec environ 40 000 cellules appartenant à plus de 100 types cellulaires, cette dernière est la structure la plus complexe du lobe optique. 800 “colonnes” représentent les unités fonctionnelles de la medulla qui traitent les informations visuelles provenant d’un point de l’espace d’une manière rétinotopique. L’organisation rétinotopique de la medulla est en quelque sorte préservée dans le complexe lobula, où le mouvement (lobula plate) et d’autres caractéristiques visuelles discutées en détail plus tard (lobula), sont traités.
La grande majorité des neurones du lobe optique de la drosophile sont générés au cours du troisième stade larvaire. Deux structures neuroépithéliales génèrent des cellules souches neurales (neuroblastes) qui se divisent plusieurs fois, s’auto-renouvelant et générant un précurseur intermédiaire, appelé cellule mère ganglionnaire (GMC), qui ne se divise qu’une fois pour générer deux neurones. Les neuroblastes sont capables de générer de nombreux types de neurones grâce à trois mécanismes principaux. i) Configuration temporelle : onze facteurs de transcription (FT), Oaz, Hth, Opa, Erm, Ey, Hbn, Slp, Scro, D, BarHI et Tll (figure 4) sont exprimés dans une séquence temporelle étroite dans chaque neuroblaste, ce qui entraîne la formation des différentes populations neuronales. ii) Configuration spatiale : trois FT exprimées dans des sous-régions du neuroépithélium (Vsx, Optix, Rx) qui génèrent la médulla modifient l’identité des neuroblastes sans affecter la séquence temporelle pour générer localement plus de types de neurones. iii) Décision binaire du destin cellulaire pilotée par Notch : une plus grande diversité neuronale est obtenue par Notch pendant la dernière division asymétrique de la GMC. La combinaison de ces trois mécanismes, ainsi que la mort cellulaire, sont responsables de la génération d’une grande partie de la diversité neuronale observée.
Figure 4
Nous cherchons à comprendre comment ces mécanismes de spécification neuronale ont évolué et comment ils se diversifient pour générer de nouveaux types cellulaires.
Responsable :
Nikos KONSTANTINIDES
Email: nikos.konstantinides(at)ijm.fr
Phone: +33 (0) 1 57 27 81 45
Membres de l’équipe :
| Konstantina | FILIPPOPOULOU | Chercheur |
| Marianne | MALARTRE | Maître de conférence |
| Elisavet | ILIOPOULOU | Ingénieure en biologie |
| Rebekah | RAKOTONIRINA-RICQUEBOURG | PhD Student |
| Leonardo | TADINI | Master 2 |
| Félix | SIMON | Postdoctorant |
| Myrto | MITLETTON | |
| Kyriaki Niovi | RAFAILIDOU |
Konstantinides N.*,$, Holguera I. $, Rossi A.M. ,$, Escobar A., Dudragne L., Chen Y.C., Tran T., Martinez Jaimes A., Özel M.N., Simon F., Shao Z., Tsankova N.M., Fullard J.F., Walldorf U., Roussos P., Desplan C.* (2021) A comprehensive series of temporal transcription factors in the fly visual system.
Nature (accepted)
$ co-first author, * co-corresponding author
Simon F.*, Konstantinides N.* (2021) Single-cell transcriptomics in the Drosophila visual system: advances and perspectives on cell identity regulation, connectivity, and neuronal diversity evolution.
Developmental Biology 479:107-122
* co-corresponding author
Özel M.N., Jafari S., Holguera I., Chen Y.C, Benhra N., El-Danaf R.N., Kapuralin K., Malin J.A., Konstantinides N.*, Desplan C. (2021) Transient and heterogeneous neuronal types revealed by a comprehensive developmental atlas of the Drosophila optic lobe.
Nature 589(7840):88-95
* co-corresponding author
Konstantinides N.*, and Desplan C.* (2020) Neuronal differentiation strategies: insights from single-cell sequencing and machine learning.
Development 147: dev193631
* co-corresponding author
Konstantinides N.*, Kapuralin K., Fadil C., Barboza L., Satija R., Desplan C. (2018) Phenotypic convergence in the brain: distinct transcription factors regulate common terminal neuronal characters.
Cell 174(3):622-635.
* co-corresponding author
Konstantinides N.*, Degabriel S., Desplan C. (2018) Neuro-evo-devo in the single cell sequencing era.
Curr Opin Syst Biol 11:32-40.
* corresponding author
Konstantinides N.* and Averof M. (2014) A common cellular basis for muscle regeneration in arthropods and vertebrates.
Science 343(6172):788-91
* co-corresponding
Publications
Preprint
Revues
- Toni Gabaldon (IRB, Barcelona)
- Michael Perry (University of California San Diego)